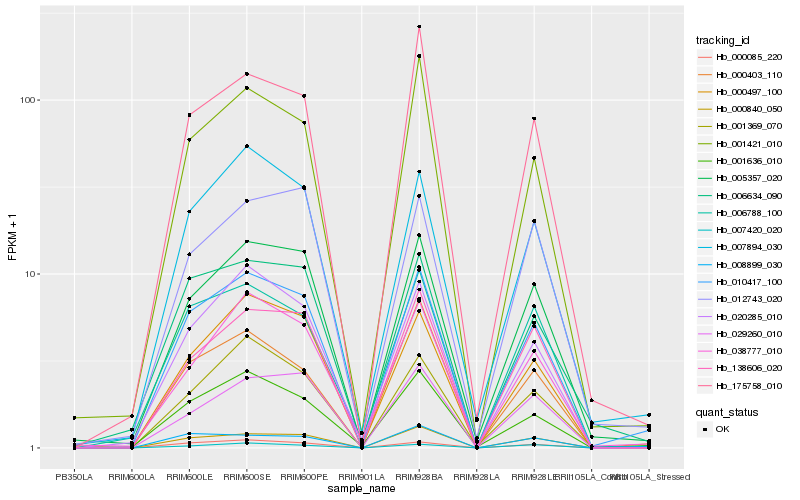
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010417_100 |
0.0 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 2 |
Hb_005357_020 |
0.079953713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007894_030 |
0.0830583508 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_001636_010 |
0.0832112782 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 5 |
Hb_000840_050 |
0.0953196019 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 6 |
Hb_000085_220 |
0.099390189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000497_100 |
0.1023702445 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 8 |
Hb_001421_010 |
0.103214857 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_020285_010 |
0.1036679755 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 10 |
Hb_006634_090 |
0.1044563483 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 11 |
Hb_038777_010 |
0.1054473043 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 12 |
Hb_001369_070 |
0.1079386685 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_006788_100 |
0.1085677193 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 14 |
Hb_175758_010 |
0.1143671598 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_138606_020 |
0.1147668698 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 16 |
Hb_000403_110 |
0.1227477098 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 17 |
Hb_008899_030 |
0.1241595141 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 18 |
Hb_012743_020 |
0.1244275238 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 19 |
Hb_029260_010 |
0.1265091382 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 20 |
Hb_007420_020 |
0.1265942757 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |