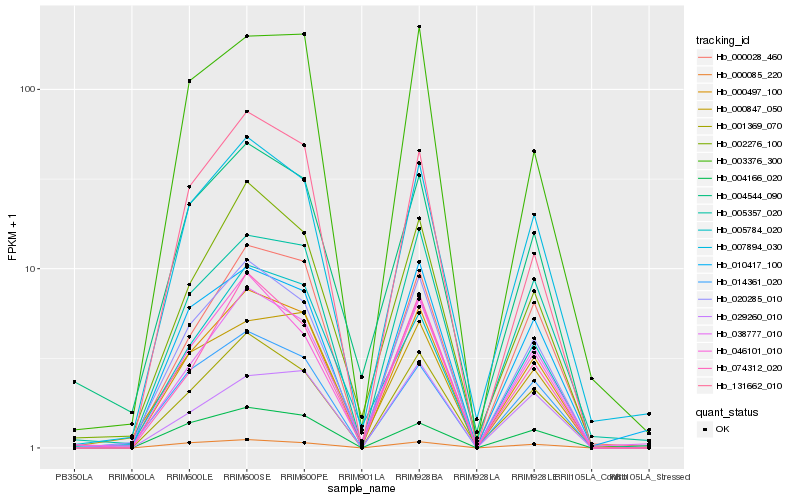
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_100 |
0.0 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 2 |
Hb_020285_010 |
0.0578381435 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 3 |
Hb_001369_070 |
0.0734191293 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000028_460 |
0.0785509502 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |
| 5 |
Hb_007894_030 |
0.0828079212 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 6 |
Hb_038777_010 |
0.0857110088 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 7 |
Hb_131662_010 |
0.0935520417 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 8 |
Hb_005357_020 |
0.0945165071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_014361_020 |
0.0950429328 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 10 |
Hb_000085_220 |
0.0950717362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_046101_010 |
0.0950814998 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 12 |
Hb_002276_100 |
0.0959838548 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 13 |
Hb_074312_020 |
0.0967675149 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_004166_020 |
0.098961028 |
- |
- |
- |
| 15 |
Hb_010417_100 |
0.1023702445 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 16 |
Hb_000847_050 |
0.1087183467 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 17 |
Hb_005784_020 |
0.1088517878 |
- |
- |
Protein MKS1, putative [Ricinus communis] |
| 18 |
Hb_004544_090 |
0.112192072 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_003376_300 |
0.1122851527 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 20 |
Hb_029260_010 |
0.11438518 |
- |
- |
nitrate transporter, putative [Ricinus communis] |