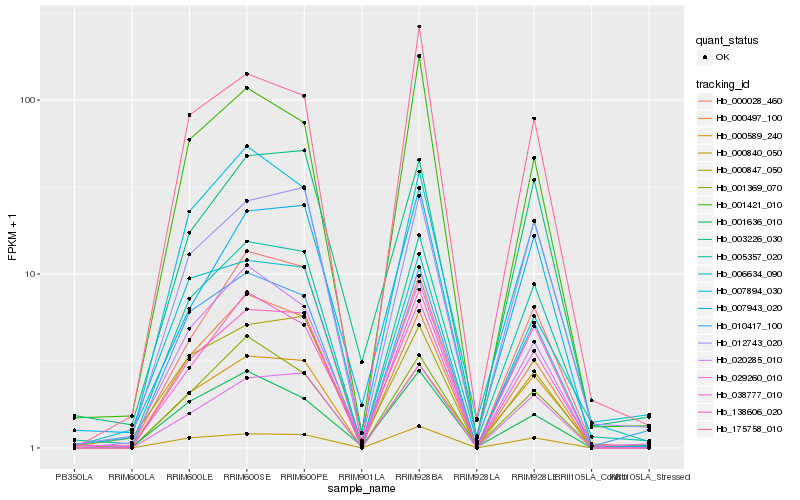
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005357_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012743_020 |
0.0731510052 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_010417_100 |
0.079953713 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 4 |
Hb_038777_010 |
0.0827622458 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 5 |
Hb_000589_240 |
0.0844529095 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_138606_020 |
0.0885667039 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 7 |
Hb_029260_010 |
0.0890050376 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 8 |
Hb_007894_030 |
0.0901923039 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000028_460 |
0.0929084594 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |
| 10 |
Hb_000497_100 |
0.0945165071 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 11 |
Hb_003226_030 |
0.1002942017 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 12 |
Hb_000840_050 |
0.1034811685 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 13 |
Hb_001369_070 |
0.1070300955 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_006634_090 |
0.1106528881 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 15 |
Hb_020285_010 |
0.1116164109 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 16 |
Hb_000847_050 |
0.1129469998 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 17 |
Hb_007943_020 |
0.115020781 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001421_010 |
0.1166126198 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001636_010 |
0.1170428059 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 20 |
Hb_175758_010 |
0.1238852866 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |