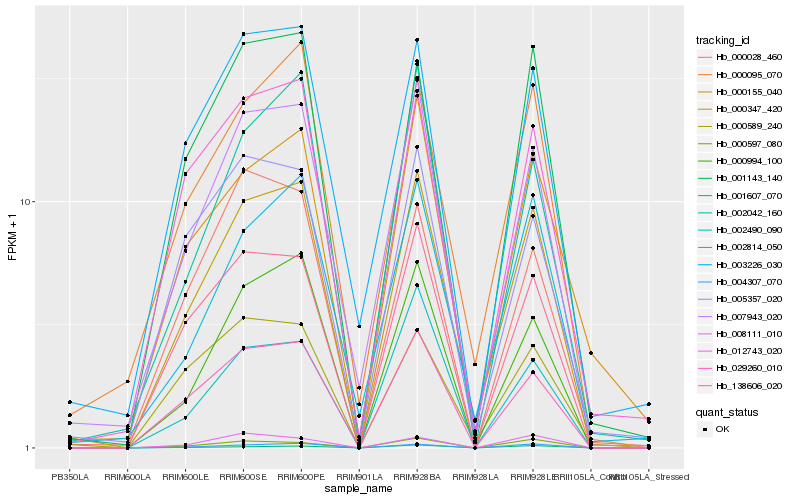
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_420 |
0.0 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 2 |
Hb_007943_020 |
0.0914902985 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_029260_010 |
0.0936059699 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_004307_070 |
0.1001514028 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_138606_020 |
0.1020659752 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 6 |
Hb_000095_070 |
0.107990956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000994_100 |
0.1081337041 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 8 |
Hb_001143_140 |
0.1105077624 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_012743_020 |
0.1175519111 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 10 |
Hb_002814_050 |
0.119065037 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 11 |
Hb_000589_240 |
0.1239355257 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_003226_030 |
0.1250998575 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 13 |
Hb_000597_080 |
0.1275587999 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_005357_020 |
0.1294123446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002042_160 |
0.1297323876 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_000155_040 |
0.1323240675 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 17 |
Hb_001607_070 |
0.139892549 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 18 |
Hb_002490_090 |
0.1419587108 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 19 |
Hb_008111_010 |
0.1432518927 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 20 |
Hb_000028_460 |
0.1436337441 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |