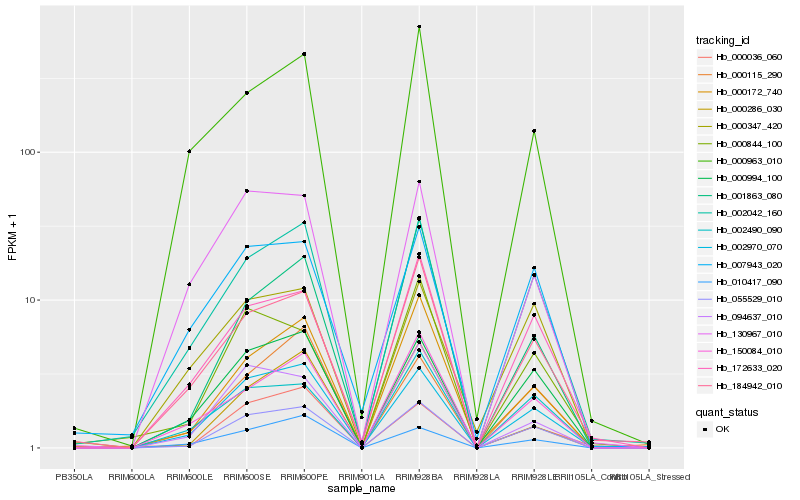
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055529_010 |
0.0 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 2 |
Hb_000994_100 |
0.0881307844 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 3 |
Hb_002970_070 |
0.0919505114 |
- |
- |
multidrug/pheromone exporter protein [Hevea brasiliensis] |
| 4 |
Hb_002042_160 |
0.1049028171 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 5 |
Hb_172633_020 |
0.1312170573 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 [Populus euphratica] |
| 6 |
Hb_002490_090 |
0.1352245428 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 7 |
Hb_000172_740 |
0.1375167094 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 8 |
Hb_184942_010 |
0.1417256778 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000036_060 |
0.1425965429 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 10 |
Hb_150084_010 |
0.1451466494 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 11 |
Hb_000115_290 |
0.1531885386 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 12 |
Hb_007943_020 |
0.1551511054 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_094637_010 |
0.1561535918 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 14 |
Hb_000844_100 |
0.156566181 |
- |
- |
hypothetical protein JCGZ_04351 [Jatropha curcas] |
| 15 |
Hb_010417_090 |
0.1608158497 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 16 |
Hb_130967_010 |
0.1608620087 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000347_420 |
0.1627545581 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 18 |
Hb_000286_030 |
0.1652587487 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 19 |
Hb_001863_080 |
0.1654940834 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 20 |
Hb_000963_010 |
0.1657109084 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |