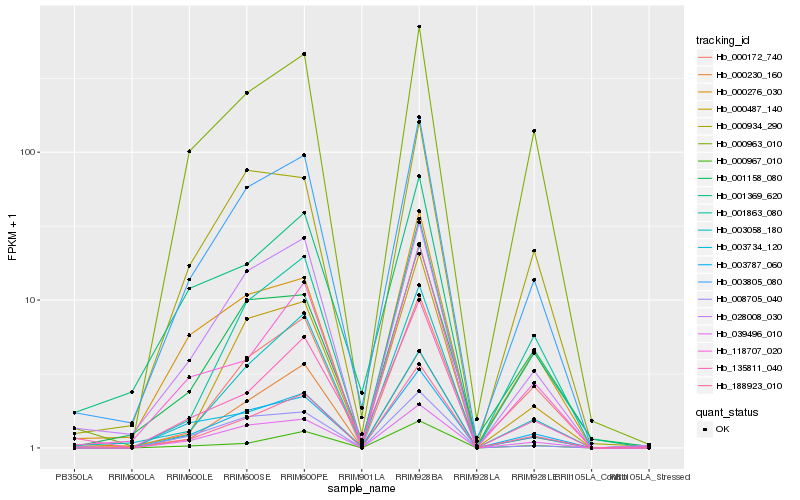
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003805_080 |
0.0 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 2 |
Hb_003787_060 |
0.058077985 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 3 |
Hb_028008_030 |
0.0790272729 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 4 |
Hb_039496_010 |
0.0814941458 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 5 |
Hb_188923_010 |
0.0827683913 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 6 |
Hb_000230_160 |
0.0931851989 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003058_180 |
0.1002865839 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 8 |
Hb_000487_140 |
0.1051073056 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000934_290 |
0.1063058711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000276_030 |
0.111652002 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_000963_010 |
0.113634605 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 12 |
Hb_000172_740 |
0.1137866409 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 13 |
Hb_001158_080 |
0.1150818376 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 14 |
Hb_008705_040 |
0.1160055758 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 15 |
Hb_003734_120 |
0.1183365752 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001863_080 |
0.1207916522 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 17 |
Hb_001369_620 |
0.1209921042 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 18 |
Hb_135811_040 |
0.1247511698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000967_010 |
0.1283571848 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 20 |
Hb_118707_020 |
0.1294731965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |