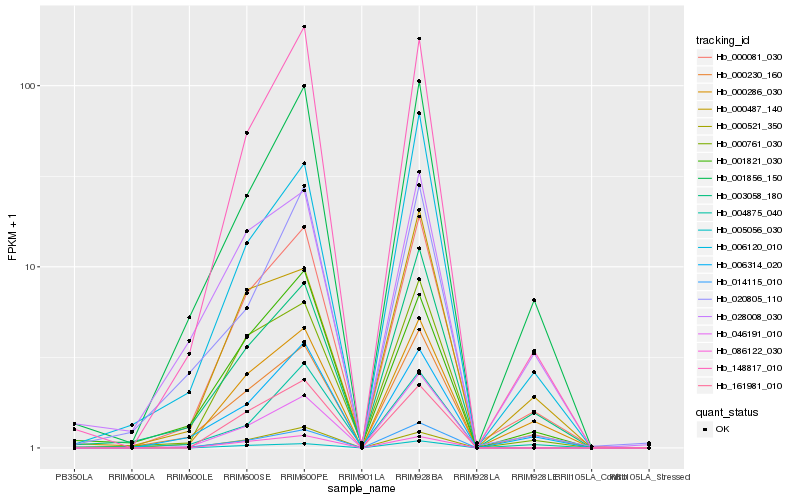
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000081_030 |
0.0 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 2 |
Hb_000230_160 |
0.0727014683 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000286_030 |
0.0801275802 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 4 |
Hb_000761_030 |
0.0863880663 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 5 |
Hb_148817_010 |
0.0914805495 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 6 |
Hb_001856_150 |
0.0918287179 |
- |
- |
- |
| 7 |
Hb_006314_020 |
0.0978210318 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 8 |
Hb_003058_180 |
0.1033209603 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 9 |
Hb_014115_010 |
0.1106673412 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001821_030 |
0.1238170777 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 11 |
Hb_006120_010 |
0.1256222536 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 12 |
Hb_161981_010 |
0.1276184214 |
- |
- |
brassinosteroid insensitive 1 -like protein [Gossypium arboreum] |
| 13 |
Hb_004875_040 |
0.1281533053 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 14 |
Hb_000521_350 |
0.1288348708 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_046191_010 |
0.1299117427 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_028008_030 |
0.1309449883 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 17 |
Hb_086122_030 |
0.1340576238 |
- |
- |
D-glycerate 3-kinase, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000487_140 |
0.1365778768 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_020805_110 |
0.1367381748 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 20 |
Hb_005056_030 |
0.1371265834 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |