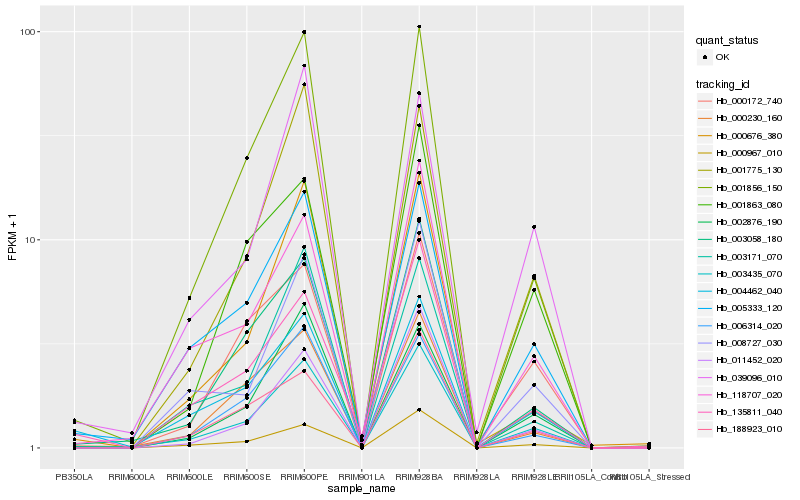
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003435_070 |
0.0 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 2 |
Hb_005333_120 |
0.083354325 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 3 |
Hb_000967_010 |
0.0885382518 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 4 |
Hb_001856_150 |
0.0910807038 |
- |
- |
- |
| 5 |
Hb_118707_020 |
0.0992882357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003058_180 |
0.1022472172 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 7 |
Hb_008727_030 |
0.1053935983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004462_040 |
0.1056586525 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000230_160 |
0.1108817126 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001775_130 |
0.1151785213 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 11 |
Hb_000172_740 |
0.1155392385 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 12 |
Hb_002876_190 |
0.1158639873 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 13 |
Hb_188923_010 |
0.1183637067 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 14 |
Hb_006314_020 |
0.123944066 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 15 |
Hb_011452_020 |
0.1254843789 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |
| 16 |
Hb_039096_010 |
0.1288569891 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_001863_080 |
0.1303118533 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 18 |
Hb_000676_380 |
0.1311532193 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 19 |
Hb_003171_070 |
0.1311717675 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 20 |
Hb_135811_040 |
0.1322189928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |