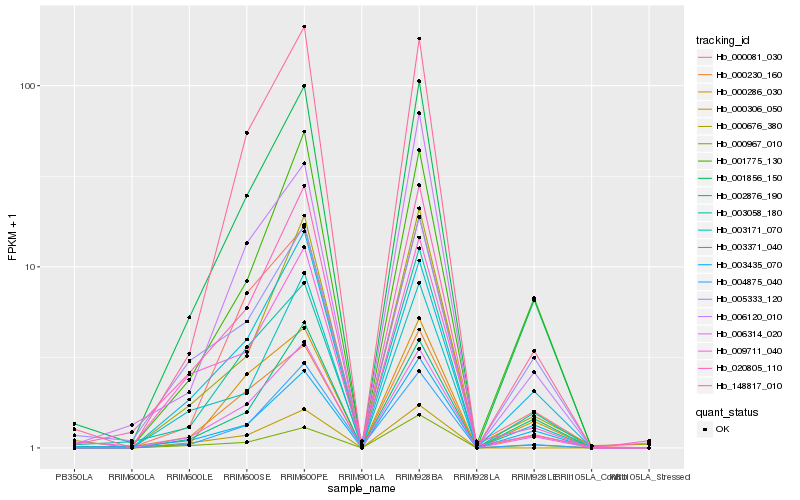
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001856_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_006314_020 |
0.0531484879 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 3 |
Hb_000230_160 |
0.0745275889 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003435_070 |
0.0910807038 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000081_030 |
0.0918287179 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 6 |
Hb_003171_070 |
0.0939377813 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 7 |
Hb_004875_040 |
0.0942594163 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 8 |
Hb_000676_380 |
0.0957483006 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 9 |
Hb_003058_180 |
0.0982205392 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 10 |
Hb_148817_010 |
0.0988526051 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 11 |
Hb_020805_110 |
0.1039453758 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 12 |
Hb_003371_040 |
0.1044300552 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 13 |
Hb_001775_130 |
0.1055882818 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 14 |
Hb_000286_030 |
0.1069108048 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 15 |
Hb_002876_190 |
0.1074646524 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 16 |
Hb_005333_120 |
0.1089486082 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 17 |
Hb_009711_040 |
0.1140005415 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 18 |
Hb_000967_010 |
0.1206327935 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 19 |
Hb_000306_050 |
0.1279180571 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_006120_010 |
0.128521976 |
- |
- |
invertase inhibitor [Manihot esculenta] |