| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
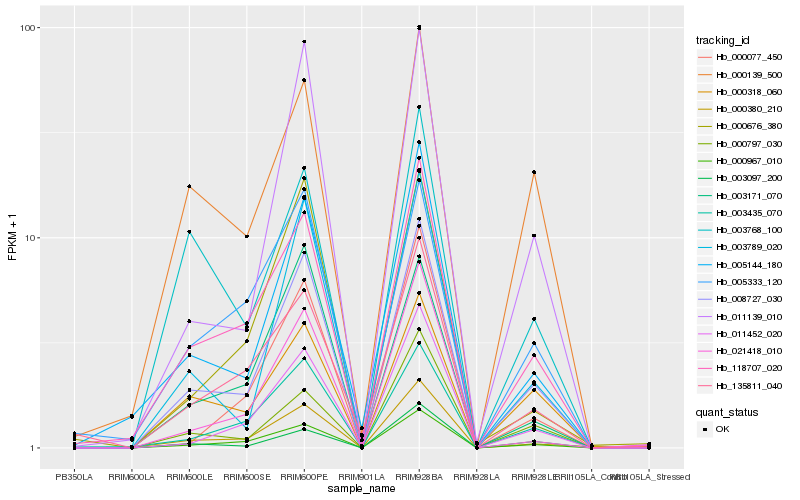
Hb_008727_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000967_010 |
0.082575984 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 3 |
Hb_118707_020 |
0.0906750548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003435_070 |
0.1053935983 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 5 |
Hb_003789_020 |
0.1062876186 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 6 |
Hb_005144_180 |
0.1093436139 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 7 |
Hb_011139_010 |
0.1119327418 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000318_060 |
0.1143430051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011452_020 |
0.1149006173 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_000380_210 |
0.1194257633 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 11 |
Hb_003097_200 |
0.1297211995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000139_500 |
0.130660621 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0286901 [Prunus mume] |
| 13 |
Hb_003171_070 |
0.134647162 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 14 |
Hb_000797_030 |
0.1356077647 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 15 |
Hb_005333_120 |
0.1363280389 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 16 |
Hb_021418_010 |
0.13899419 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 17 |
Hb_000676_380 |
0.1407433081 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 18 |
Hb_135811_040 |
0.1417704354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003768_100 |
0.1429749227 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 20 |
Hb_000077_450 |
0.1444062532 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |