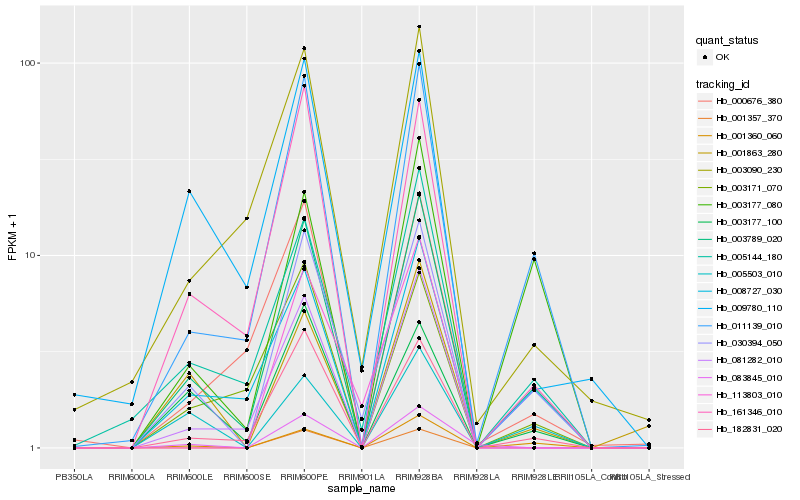
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030394_050 |
0.0 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 2 |
Hb_003789_020 |
0.108807589 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 3 |
Hb_011139_010 |
0.1351146846 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_182831_020 |
0.1446206917 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_001360_060 |
0.1511057929 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 6 |
Hb_161346_010 |
0.1644472651 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 7 |
Hb_005144_180 |
0.1684506582 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 8 |
Hb_008727_030 |
0.172532463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003177_100 |
0.1737125995 |
- |
- |
PREDICTED: auxin-induced protein 6B [Jatropha curcas] |
| 10 |
Hb_005503_010 |
0.1833780029 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 11 |
Hb_009780_110 |
0.1861064185 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 12 |
Hb_081282_010 |
0.1970696971 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 13 |
Hb_003090_230 |
0.1994382447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003177_080 |
0.2012531127 |
- |
- |
hypothetical protein L484_016032 [Morus notabilis] |
| 15 |
Hb_003171_070 |
0.2018186542 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 16 |
Hb_083845_010 |
0.2019157778 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 17 |
Hb_113803_010 |
0.2049563198 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000676_380 |
0.2054547087 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 19 |
Hb_001863_280 |
0.207161522 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 20 |
Hb_001357_370 |
0.2111967524 |
- |
- |
- |