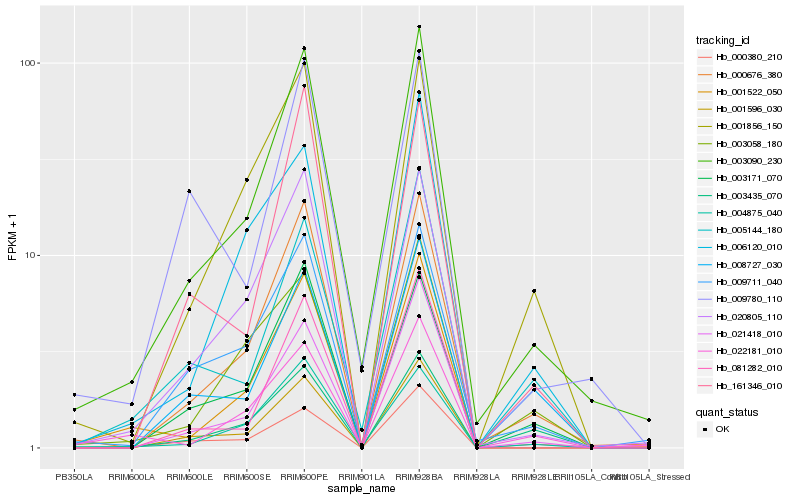
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003090_230 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000676_380 |
0.0776606865 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 3 |
Hb_020805_110 |
0.0972825237 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_009711_040 |
0.1138636792 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 5 |
Hb_021418_010 |
0.1156416559 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 6 |
Hb_004875_040 |
0.1201148302 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 7 |
Hb_001596_030 |
0.121205308 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 8 |
Hb_001522_050 |
0.1218403486 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 9 |
Hb_000380_210 |
0.1295709172 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 10 |
Hb_005144_180 |
0.1301322815 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 11 |
Hb_003171_070 |
0.1343042172 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 12 |
Hb_001856_150 |
0.1347418119 |
- |
- |
- |
| 13 |
Hb_081282_010 |
0.1349345286 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 14 |
Hb_003058_180 |
0.1424532473 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 15 |
Hb_009780_110 |
0.1438442046 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 16 |
Hb_022181_010 |
0.1459126233 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 17 |
Hb_006120_010 |
0.1464465936 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 18 |
Hb_003435_070 |
0.1479016051 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 19 |
Hb_161346_010 |
0.1490802517 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 20 |
Hb_008727_030 |
0.1517075399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |