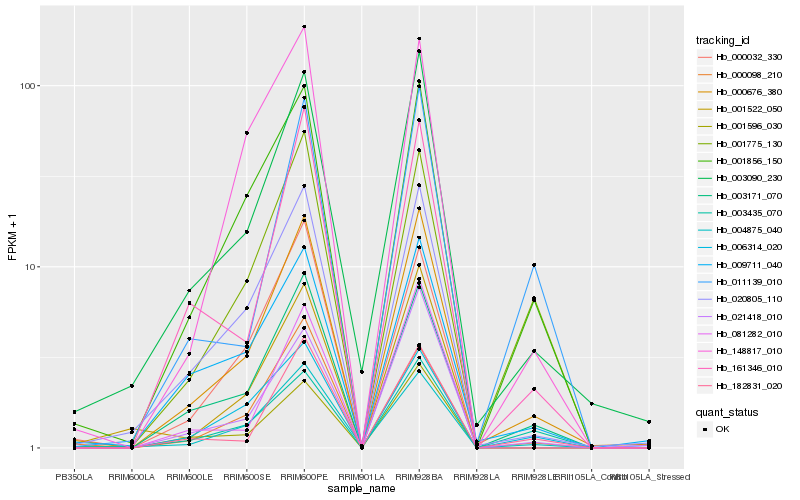
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_380 |
0.0 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 2 |
Hb_003090_230 |
0.0776606865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_020805_110 |
0.0873445868 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_003171_070 |
0.0906181137 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 5 |
Hb_001856_150 |
0.0957483006 |
- |
- |
- |
| 6 |
Hb_004875_040 |
0.1036415361 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 7 |
Hb_009711_040 |
0.1056539701 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 8 |
Hb_182831_020 |
0.1205342837 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 9 |
Hb_161346_010 |
0.1210930676 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 10 |
Hb_021418_010 |
0.1219045029 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 11 |
Hb_001596_030 |
0.1220635962 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 12 |
Hb_006314_020 |
0.1271803738 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 13 |
Hb_000098_210 |
0.1289188662 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 14 |
Hb_001775_130 |
0.1304003878 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 15 |
Hb_003435_070 |
0.1311532193 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 16 |
Hb_148817_010 |
0.1331245617 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 17 |
Hb_001522_050 |
0.1349062853 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 18 |
Hb_081282_010 |
0.1363594705 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 19 |
Hb_000032_330 |
0.1370516023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011139_010 |
0.1393228382 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |