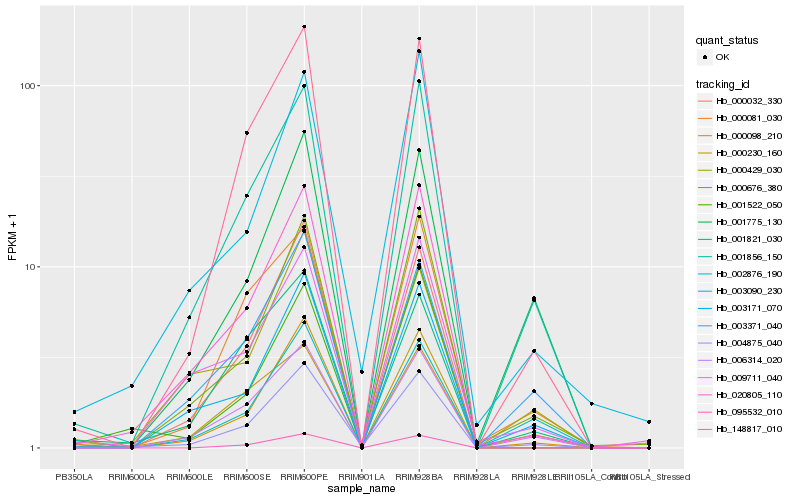
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004875_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 2 |
Hb_020805_110 |
0.0912600032 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 3 |
Hb_148817_010 |
0.0934316582 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 4 |
Hb_001856_150 |
0.0942594163 |
- |
- |
- |
| 5 |
Hb_000098_210 |
0.096870345 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 6 |
Hb_000676_380 |
0.1036415361 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 7 |
Hb_006314_020 |
0.1071741721 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 8 |
Hb_000032_330 |
0.1071911422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003171_070 |
0.1133673697 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 10 |
Hb_003090_230 |
0.1201148302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001821_030 |
0.1234513944 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 12 |
Hb_001775_130 |
0.1264163234 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 13 |
Hb_000081_030 |
0.1281533053 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 14 |
Hb_095532_010 |
0.1300614238 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 15 |
Hb_003371_040 |
0.1310987224 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 16 |
Hb_002876_190 |
0.1327509208 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 17 |
Hb_000230_160 |
0.1332408158 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009711_040 |
0.1382189022 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 19 |
Hb_000429_030 |
0.1415406809 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001522_050 |
0.1440254691 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |