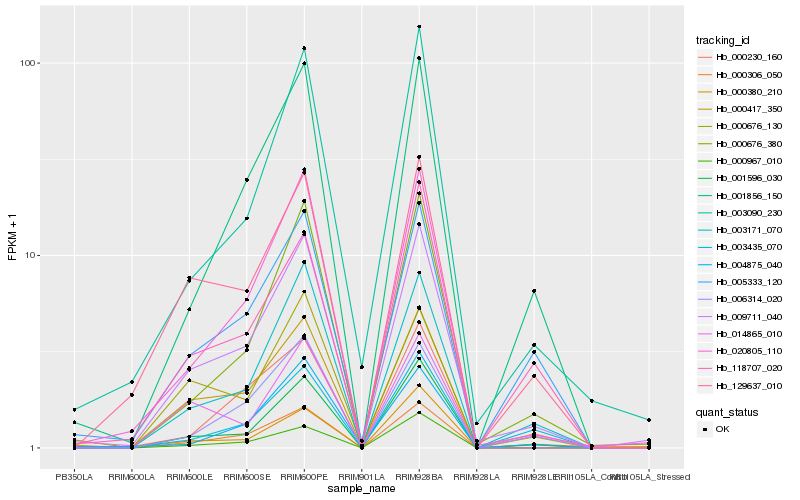
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009711_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 2 |
Hb_000417_350 |
0.0779573239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_020805_110 |
0.088901136 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_000676_380 |
0.1056539701 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 5 |
Hb_003171_070 |
0.1101275714 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 6 |
Hb_000306_050 |
0.110882394 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_003090_230 |
0.1138636792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001856_150 |
0.1140005415 |
- |
- |
- |
| 9 |
Hb_129637_010 |
0.1224448564 |
- |
- |
PREDICTED: alpha-mannosidase isoform X1 [Populus euphratica] |
| 10 |
Hb_001596_030 |
0.1249020689 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 11 |
Hb_005333_120 |
0.1252039148 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 12 |
Hb_006314_020 |
0.1285325735 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 13 |
Hb_000230_160 |
0.1357905207 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004875_040 |
0.1382189022 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 15 |
Hb_000380_210 |
0.141161245 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 16 |
Hb_000967_010 |
0.1437530497 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 17 |
Hb_118707_020 |
0.1444239225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003435_070 |
0.1455847688 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000676_130 |
0.1455910359 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 20 |
Hb_014865_010 |
0.1474929477 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |