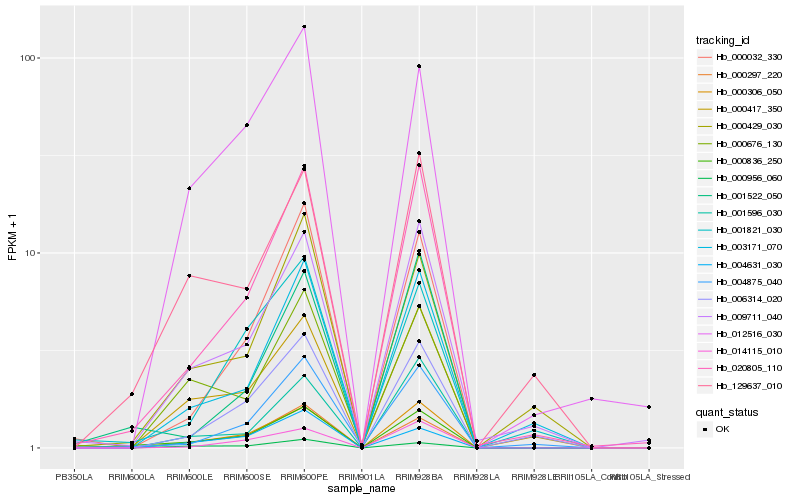
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_250 |
0.0 |
- |
- |
hypothetical protein VITISV_022299 [Vitis vinifera] |
| 2 |
Hb_020805_110 |
0.1147882051 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 3 |
Hb_000306_050 |
0.1152852208 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000297_220 |
0.1227014351 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 5 |
Hb_129637_010 |
0.1437005394 |
- |
- |
PREDICTED: alpha-mannosidase isoform X1 [Populus euphratica] |
| 6 |
Hb_000417_350 |
0.145090204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009711_040 |
0.1479056188 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 8 |
Hb_012516_030 |
0.1489597762 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 9 |
Hb_000956_060 |
0.1530877236 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000032_330 |
0.1582320098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000676_130 |
0.1582615924 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 12 |
Hb_004875_040 |
0.1603400906 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 13 |
Hb_006314_020 |
0.1620465798 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 14 |
Hb_001821_030 |
0.1652586214 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 15 |
Hb_004631_030 |
0.1652726965 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 16 |
Hb_003171_070 |
0.1655741373 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 17 |
Hb_014115_010 |
0.1657818266 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000429_030 |
0.1658735265 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 19 |
Hb_001522_050 |
0.1664290811 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 20 |
Hb_001596_030 |
0.1674604044 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |