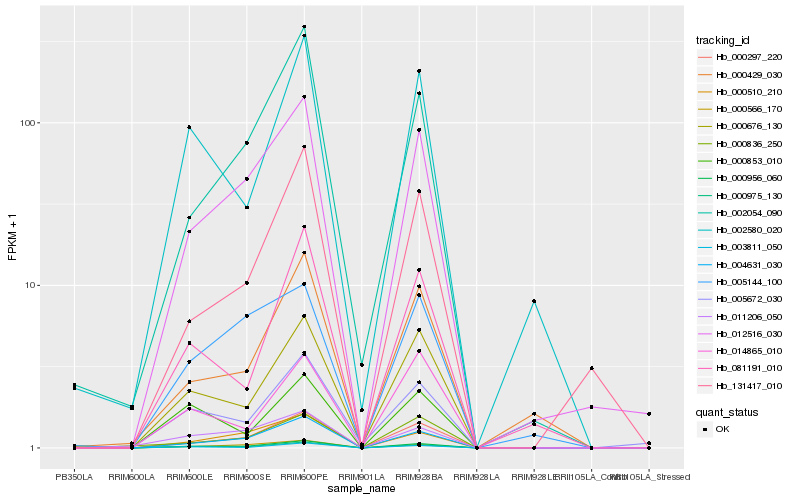
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000956_060 |
0.0 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000975_130 |
0.057788886 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 3 |
Hb_000297_220 |
0.0778688286 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 4 |
Hb_004631_030 |
0.0796671914 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 5 |
Hb_012516_030 |
0.0852382852 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 6 |
Hb_003811_050 |
0.0980826778 |
- |
- |
hypothetical protein JCGZ_01310 [Jatropha curcas] |
| 7 |
Hb_000676_130 |
0.1019745957 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 8 |
Hb_000566_170 |
0.1192505927 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 9 |
Hb_005672_030 |
0.1223810664 |
- |
- |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 10 |
Hb_000510_210 |
0.1303612874 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_002054_090 |
0.1424409208 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 12 |
Hb_011206_050 |
0.1434009426 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 13 |
Hb_002580_020 |
0.1441828715 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 14 |
Hb_131417_010 |
0.1495033111 |
- |
- |
hypothetical protein POPTR_0011s066602g, partial [Populus trichocarpa] |
| 15 |
Hb_000429_030 |
0.1498197127 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 16 |
Hb_081191_010 |
0.1524319501 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 17 |
Hb_000836_250 |
0.1530877236 |
- |
- |
hypothetical protein VITISV_022299 [Vitis vinifera] |
| 18 |
Hb_000853_010 |
0.153504541 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 19 |
Hb_014865_010 |
0.154905236 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 20 |
Hb_005144_100 |
0.1583142436 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |