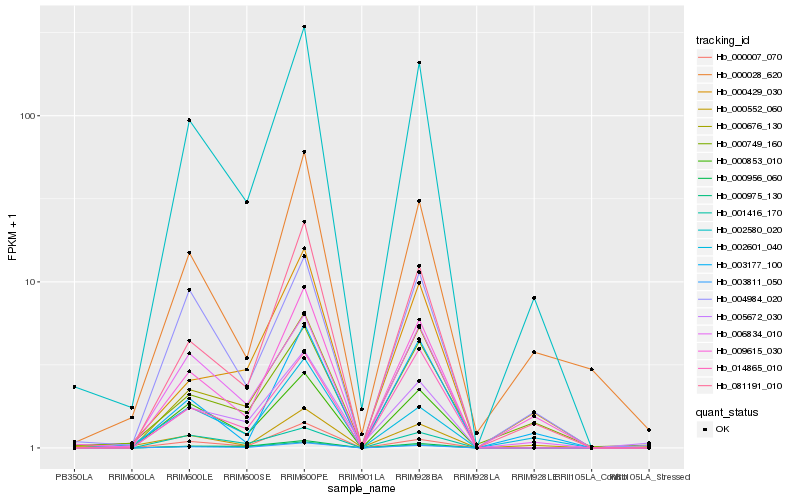
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002580_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 2 |
Hb_009615_030 |
0.0771861313 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_081191_010 |
0.0873822684 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 4 |
Hb_003811_050 |
0.0979604044 |
- |
- |
hypothetical protein JCGZ_01310 [Jatropha curcas] |
| 5 |
Hb_000676_130 |
0.1078991491 |
- |
- |
PREDICTED: peroxidase 72-like [Nelumbo nucifera] |
| 6 |
Hb_006834_010 |
0.1117506328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000853_010 |
0.1185599361 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 8 |
Hb_004984_020 |
0.1274047319 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 9 |
Hb_000975_130 |
0.1309587471 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 10 |
Hb_000429_030 |
0.1326209911 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_003177_100 |
0.1339401365 |
- |
- |
PREDICTED: auxin-induced protein 6B [Jatropha curcas] |
| 12 |
Hb_000028_620 |
0.1357047755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000749_160 |
0.1379190703 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 14 |
Hb_005672_030 |
0.14063094 |
- |
- |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 15 |
Hb_014865_010 |
0.1415385291 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 16 |
Hb_000956_060 |
0.1441828715 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000552_060 |
0.1472207941 |
- |
- |
PREDICTED: theobromine synthase 2-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_000007_070 |
0.1486190439 |
- |
- |
hypothetical protein POPTR_0008s11290g [Populus trichocarpa] |
| 19 |
Hb_001416_170 |
0.1565883683 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_002601_040 |
0.1584756164 |
- |
- |
hypothetical protein JCGZ_06197 [Jatropha curcas] |