| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
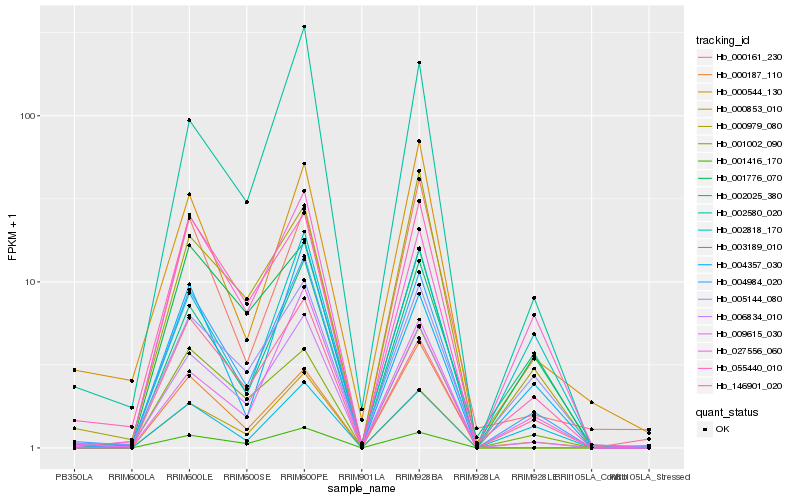
Hb_004984_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 2 |
Hb_006834_010 |
0.08796205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_055440_010 |
0.1175253502 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_000853_010 |
0.1202824804 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 5 |
Hb_002580_020 |
0.1274047319 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 6 |
Hb_005144_080 |
0.127428584 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 7 |
Hb_001416_170 |
0.1274947359 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_027556_060 |
0.1323156588 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 9 |
Hb_146901_020 |
0.1337135749 |
- |
- |
- |
| 10 |
Hb_004357_030 |
0.134914346 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_001002_090 |
0.146815072 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 12 |
Hb_003189_010 |
0.1469806251 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 13 |
Hb_000187_110 |
0.1478898799 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 14 |
Hb_000544_130 |
0.1483394677 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 15 |
Hb_002025_380 |
0.1493623059 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 16 |
Hb_001776_070 |
0.1495184092 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 17 |
Hb_000161_230 |
0.1515315879 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 18 |
Hb_009615_030 |
0.1517190185 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000979_080 |
0.15304227 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 20 |
Hb_002818_170 |
0.1545497014 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |