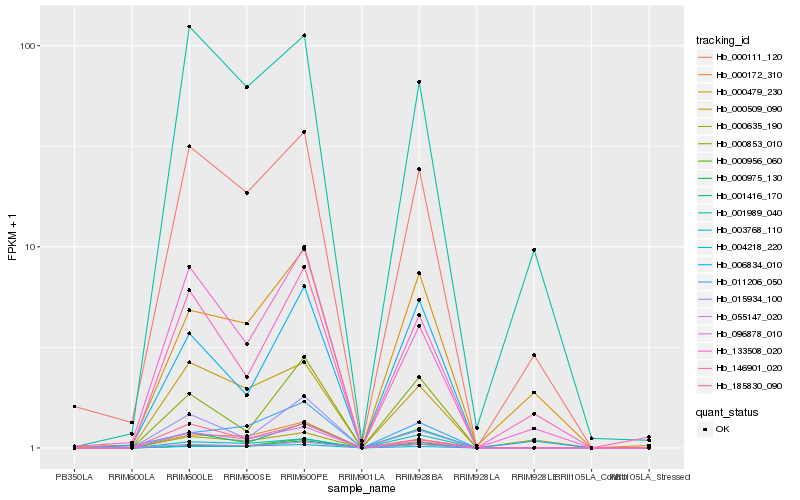
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055147_020 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 2 |
Hb_001416_170 |
0.0932461483 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_000172_310 |
0.1189912074 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 4 |
Hb_000975_130 |
0.125188328 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 5 |
Hb_004218_220 |
0.1320560409 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_096878_010 |
0.13229052 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 7 |
Hb_000111_120 |
0.1327586026 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000853_010 |
0.1380801716 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 9 |
Hb_000509_090 |
0.1407683553 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_006834_010 |
0.1451740372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_015934_100 |
0.1461553035 |
- |
- |
phospholipid/glycerol acyltransferase family protein [Populus trichocarpa] |
| 12 |
Hb_133508_020 |
0.146658434 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 13 |
Hb_185830_090 |
0.1574833234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000635_190 |
0.1606878605 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 15 |
Hb_146901_020 |
0.1630115712 |
- |
- |
- |
| 16 |
Hb_011206_050 |
0.1645434766 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 17 |
Hb_001989_040 |
0.165037003 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 18 |
Hb_003768_110 |
0.1657471796 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 19 |
Hb_000479_230 |
0.1665048521 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 20 |
Hb_000956_060 |
0.1671711578 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |