| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
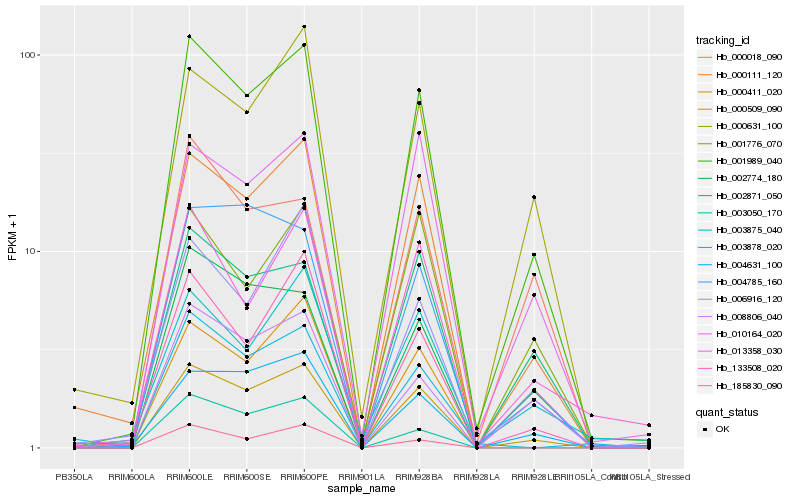
Hb_001989_040 |
0.0 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 2 |
Hb_000509_090 |
0.0463323939 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 3 |
Hb_000111_120 |
0.0885190891 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_008806_040 |
0.1208918648 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 5 |
Hb_002774_180 |
0.1238353542 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_010164_020 |
0.1255280052 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 7 |
Hb_004631_100 |
0.1268576785 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 8 |
Hb_133508_020 |
0.1269954898 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 9 |
Hb_002871_050 |
0.127371784 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 10 |
Hb_000631_100 |
0.1304724833 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 11 |
Hb_013358_030 |
0.1330504279 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 12 |
Hb_001776_070 |
0.1357871772 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 13 |
Hb_000411_020 |
0.1358035534 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_003875_040 |
0.1374593116 |
- |
- |
PREDICTED: endoglucanase 11-like [Jatropha curcas] |
| 15 |
Hb_006916_120 |
0.1405015286 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 16 |
Hb_000018_090 |
0.1455021484 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_003878_020 |
0.14628488 |
- |
- |
Epidermis-specific secreted glycoprotein EP1 precursor, putative [Ricinus communis] |
| 18 |
Hb_185830_090 |
0.1486559506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003050_170 |
0.1504431615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004785_160 |
0.1513483525 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |