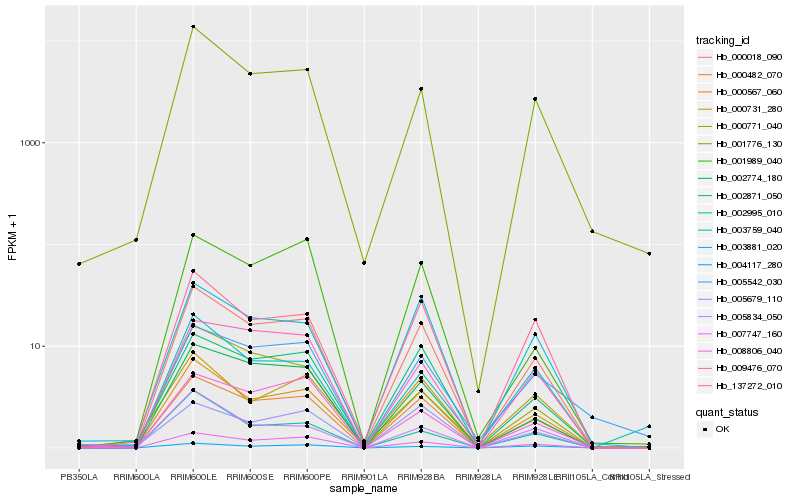
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000018_090 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000482_070 |
0.0658058395 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 3 |
Hb_000567_060 |
0.089457844 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 4 |
Hb_002871_050 |
0.0980454811 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 5 |
Hb_002774_180 |
0.1031437364 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_137272_010 |
0.1053581889 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 7 |
Hb_001776_130 |
0.1083776579 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 8 |
Hb_003881_020 |
0.1201336529 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 9 |
Hb_007747_160 |
0.1266455953 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 10 |
Hb_009476_070 |
0.1333107553 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 11 |
Hb_002995_010 |
0.1369795334 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 12 |
Hb_004117_280 |
0.139733317 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |
| 13 |
Hb_005834_050 |
0.1404095228 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 14 |
Hb_005679_110 |
0.1434903123 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 15 |
Hb_003759_040 |
0.1437444111 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 16 |
Hb_000731_280 |
0.1439624513 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008806_040 |
0.1441709043 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 18 |
Hb_005542_030 |
0.1448091697 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001989_040 |
0.1455021484 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 20 |
Hb_000771_040 |
0.1470152518 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |