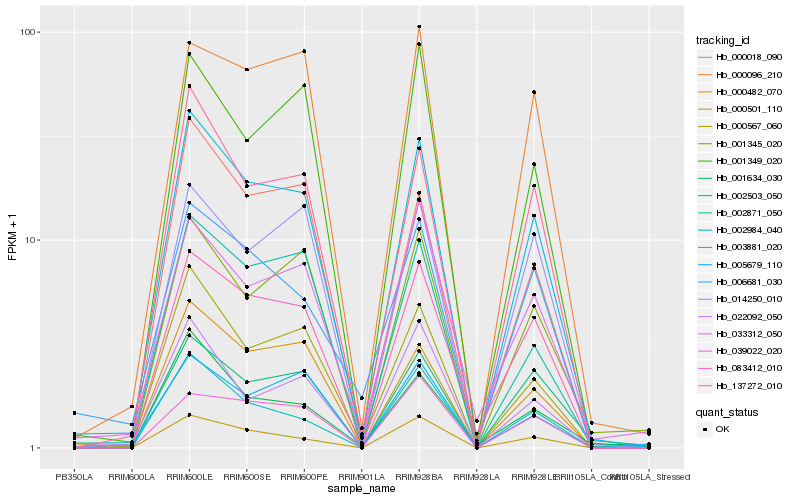
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003881_020 |
0.0 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 2 |
Hb_137272_010 |
0.0840048883 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 3 |
Hb_083412_010 |
0.0900308568 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 4 |
Hb_000567_060 |
0.0936041944 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 5 |
Hb_000482_070 |
0.1022697345 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 6 |
Hb_000501_110 |
0.1026482294 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 7 |
Hb_002984_040 |
0.1059109112 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_002871_050 |
0.1080382674 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 9 |
Hb_002503_050 |
0.1194126761 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 10 |
Hb_005679_110 |
0.1198904352 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 11 |
Hb_000018_090 |
0.1201336529 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001349_020 |
0.1213205441 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 13 |
Hb_001345_020 |
0.1223665709 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 14 |
Hb_014250_010 |
0.1241659172 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 15 |
Hb_000096_210 |
0.1295259464 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 16 |
Hb_006681_030 |
0.131962506 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_033312_050 |
0.1324404321 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 18 |
Hb_022092_050 |
0.1329903595 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 19 |
Hb_039022_020 |
0.1336749543 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 20 |
Hb_001634_030 |
0.1349846753 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |