| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
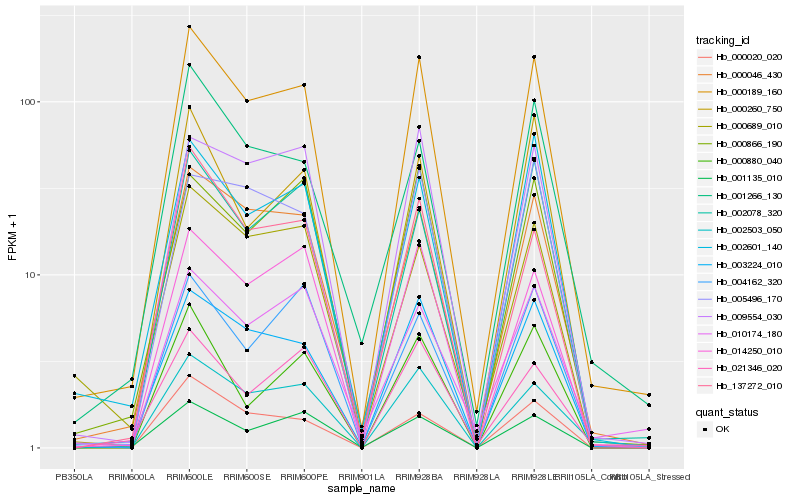
Hb_000189_160 |
0.0 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 2 |
Hb_003224_010 |
0.0754963624 |
- |
- |
- |
| 3 |
Hb_000046_430 |
0.0773015587 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 4 |
Hb_002503_050 |
0.0826434948 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 5 |
Hb_014250_010 |
0.1057949435 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 6 |
Hb_002601_140 |
0.1069748163 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 7 |
Hb_001135_010 |
0.1071273405 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 8 |
Hb_009554_030 |
0.113864768 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 9 |
Hb_002078_320 |
0.1140253298 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_000866_190 |
0.1158097297 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000020_020 |
0.1164694396 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 12 |
Hb_137272_010 |
0.117363316 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 13 |
Hb_010174_180 |
0.1188756205 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000260_750 |
0.1191947971 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 15 |
Hb_001266_130 |
0.1215147191 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 16 |
Hb_021346_020 |
0.1232343039 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 17 |
Hb_000689_010 |
0.1260578779 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_005496_170 |
0.1279988834 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 19 |
Hb_000880_040 |
0.1311953476 |
- |
- |
- |
| 20 |
Hb_004162_320 |
0.1318497724 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |