| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
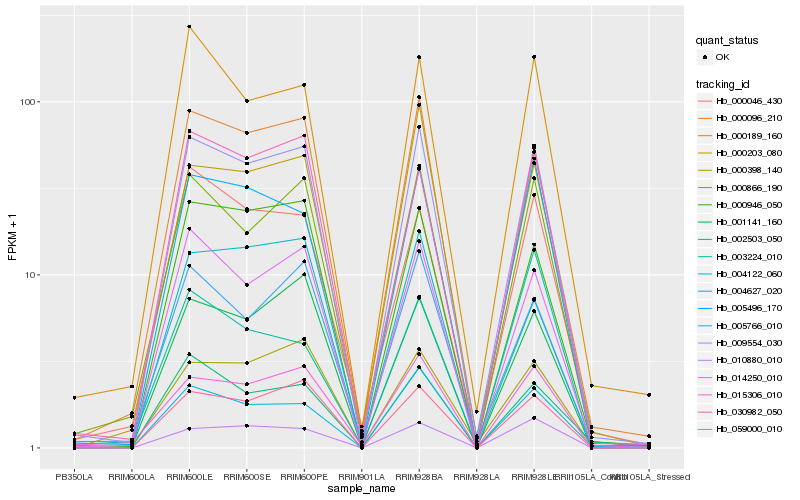
Hb_009554_030 |
0.0 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 2 |
Hb_004122_060 |
0.0710176784 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 3 |
Hb_059000_010 |
0.0768110744 |
- |
- |
hypothetical protein JCGZ_17736 [Jatropha curcas] |
| 4 |
Hb_000096_210 |
0.0793740854 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 5 |
Hb_000866_190 |
0.0796176152 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_014250_010 |
0.0813550906 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 7 |
Hb_015306_010 |
0.0978251023 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 8 |
Hb_005496_170 |
0.1016312509 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 9 |
Hb_001141_160 |
0.1050864934 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 10 |
Hb_000946_050 |
0.1064924288 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_000398_140 |
0.1080781085 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 12 |
Hb_010880_010 |
0.1084116636 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_005766_010 |
0.1089104721 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_000203_080 |
0.110889368 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 15 |
Hb_030982_050 |
0.1112687066 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 16 |
Hb_003224_010 |
0.1113009139 |
- |
- |
- |
| 17 |
Hb_002503_050 |
0.1138549704 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 18 |
Hb_000189_160 |
0.113864768 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 19 |
Hb_004627_020 |
0.1141181776 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 20 |
Hb_000046_430 |
0.1167397692 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |