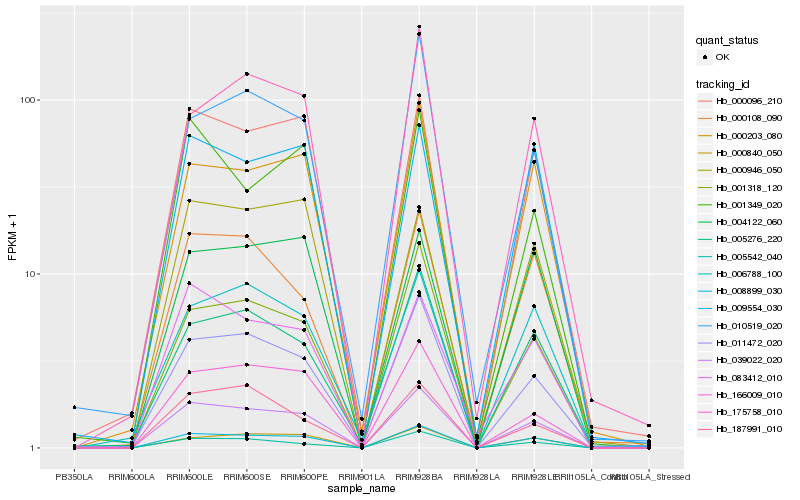
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039022_020 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 2 |
Hb_011472_020 |
0.0848384174 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 3 |
Hb_000096_210 |
0.0850093282 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 4 |
Hb_005542_040 |
0.0956043118 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 5 |
Hb_001318_120 |
0.0957015892 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 6 |
Hb_166009_010 |
0.0958700316 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 7 |
Hb_000840_050 |
0.0963756962 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 8 |
Hb_083412_010 |
0.1039011877 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_000203_080 |
0.1065710217 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 10 |
Hb_008899_030 |
0.1081555704 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 11 |
Hb_005276_220 |
0.1091388736 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 12 |
Hb_000108_090 |
0.1182800176 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 13 |
Hb_006788_100 |
0.1193360569 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 14 |
Hb_187991_010 |
0.122020289 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 15 |
Hb_175758_010 |
0.1231434869 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001349_020 |
0.1233115847 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 17 |
Hb_009554_030 |
0.1236256164 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 18 |
Hb_010519_020 |
0.1265916347 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 19 |
Hb_000946_050 |
0.1295615245 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_004122_060 |
0.1305582783 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |