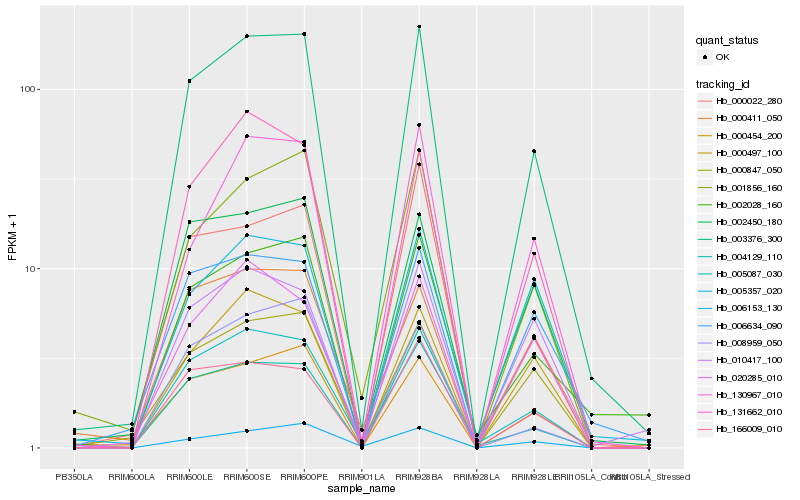
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_300 |
0.0 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 2 |
Hb_000454_200 |
0.0903343554 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 3 |
Hb_005087_030 |
0.0965747232 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 4 |
Hb_002450_180 |
0.1018244181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002028_160 |
0.1018339498 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Populus euphratica] |
| 6 |
Hb_166009_010 |
0.1022485433 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 7 |
Hb_001856_160 |
0.1072431941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000847_050 |
0.1105666295 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 9 |
Hb_000497_100 |
0.1122851527 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 10 |
Hb_131662_010 |
0.1124852328 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 11 |
Hb_006634_090 |
0.11371658 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 12 |
Hb_130967_010 |
0.1205533062 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_004129_110 |
0.1219639075 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 14 |
Hb_000022_280 |
0.1240377557 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 15 |
Hb_008959_050 |
0.1278242976 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_006153_130 |
0.128064585 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_005357_020 |
0.1289938917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010417_100 |
0.1299165339 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 19 |
Hb_020285_010 |
0.1300753365 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 20 |
Hb_000411_050 |
0.1321463531 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |