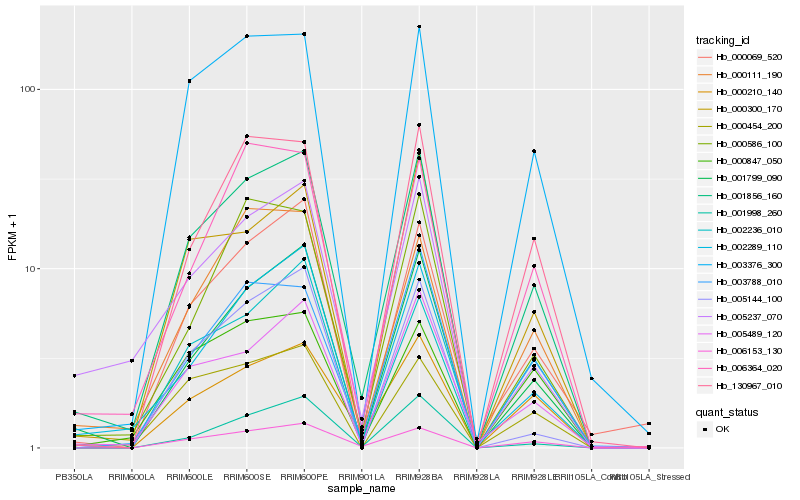
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001856_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001799_090 |
0.0891903658 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 3 |
Hb_006153_130 |
0.0930666345 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_003376_300 |
0.1072431941 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 5 |
Hb_002289_110 |
0.1082326124 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 6 |
Hb_005489_120 |
0.1089198993 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 7 |
Hb_000454_200 |
0.1123501157 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 8 |
Hb_000210_140 |
0.1145513973 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 9 |
Hb_000069_520 |
0.1176435778 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 10 |
Hb_130967_010 |
0.1188288031 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000300_170 |
0.1209161238 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 12 |
Hb_003788_010 |
0.1220761223 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 13 |
Hb_000111_190 |
0.1223319306 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 14 |
Hb_001998_260 |
0.1237063847 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_006364_020 |
0.1255700683 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 16 |
Hb_005237_070 |
0.1263217074 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 17 |
Hb_000586_100 |
0.1289324978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000847_050 |
0.1296599988 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 19 |
Hb_005144_100 |
0.1299570422 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 20 |
Hb_002236_010 |
0.1327920237 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |