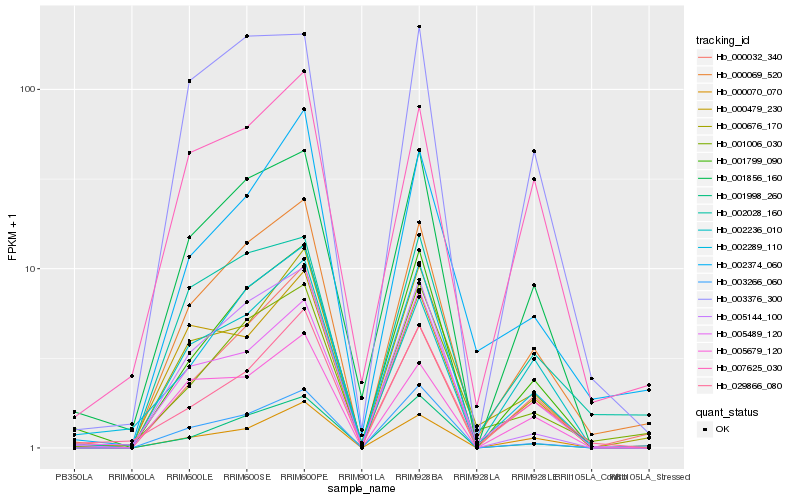
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_520 |
0.0 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 2 |
Hb_001006_030 |
0.1066185976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002028_160 |
0.1104273915 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Populus euphratica] |
| 4 |
Hb_001799_090 |
0.1119052559 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 5 |
Hb_000676_170 |
0.1141767441 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 6 |
Hb_002289_110 |
0.1148766085 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 7 |
Hb_002374_060 |
0.1174649993 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_000070_070 |
0.1176216924 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 9 |
Hb_001856_160 |
0.1176435778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002236_010 |
0.1235844232 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 11 |
Hb_003266_060 |
0.1238826716 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 12 |
Hb_000032_340 |
0.1251211541 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 13 |
Hb_005144_100 |
0.126107246 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 14 |
Hb_007625_030 |
0.1272830494 |
- |
- |
unknown [Medicago truncatula] |
| 15 |
Hb_005679_120 |
0.1372674413 |
- |
- |
hypothetical protein RCOM_0284080 [Ricinus communis] |
| 16 |
Hb_005489_120 |
0.1372994508 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 17 |
Hb_000479_230 |
0.1373243078 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 18 |
Hb_001998_260 |
0.1374277945 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_029866_080 |
0.1396153131 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 20 |
Hb_003376_300 |
0.139671236 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |