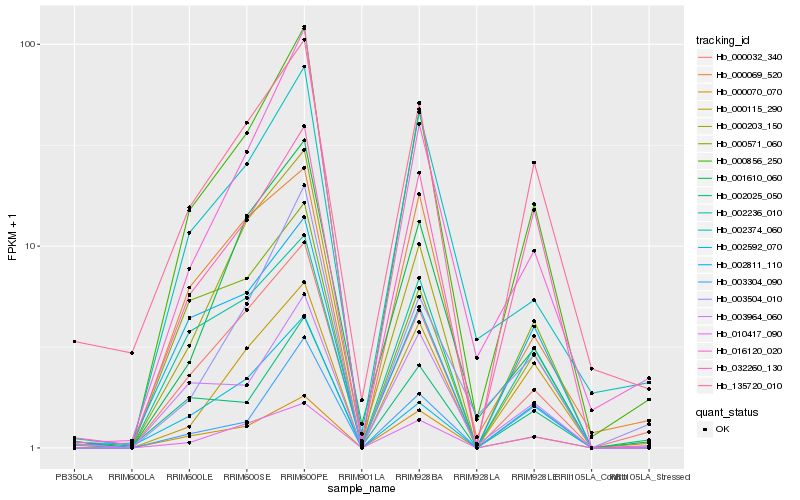
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_250 |
0.0 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000032_340 |
0.0835426341 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 3 |
Hb_016120_020 |
0.0966303199 |
- |
- |
PREDICTED: tetrahydrocannabinolic acid synthase-like [Jatropha curcas] |
| 4 |
Hb_000070_070 |
0.1086438811 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 5 |
Hb_010417_090 |
0.1145702892 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 6 |
Hb_000203_150 |
0.1214012238 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 7 |
Hb_002236_010 |
0.1251910441 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 8 |
Hb_001610_060 |
0.1276952169 |
- |
- |
hypothetical protein TRIUR3_12067 [Triticum urartu] |
| 9 |
Hb_003964_060 |
0.1318970373 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |
| 10 |
Hb_003504_010 |
0.1324708117 |
- |
- |
hypothetical protein POPTR_0013s04290g [Populus trichocarpa] |
| 11 |
Hb_002374_060 |
0.135273116 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_000571_060 |
0.1358356706 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 13 |
Hb_135720_010 |
0.138726085 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 14 |
Hb_002592_070 |
0.1393476797 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 15 |
Hb_002811_110 |
0.142313037 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 16 |
Hb_000069_520 |
0.144445396 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 17 |
Hb_000115_290 |
0.1458489358 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 18 |
Hb_002025_050 |
0.1474578657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_032260_130 |
0.1480433819 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 20 |
Hb_003304_090 |
0.1510825091 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |