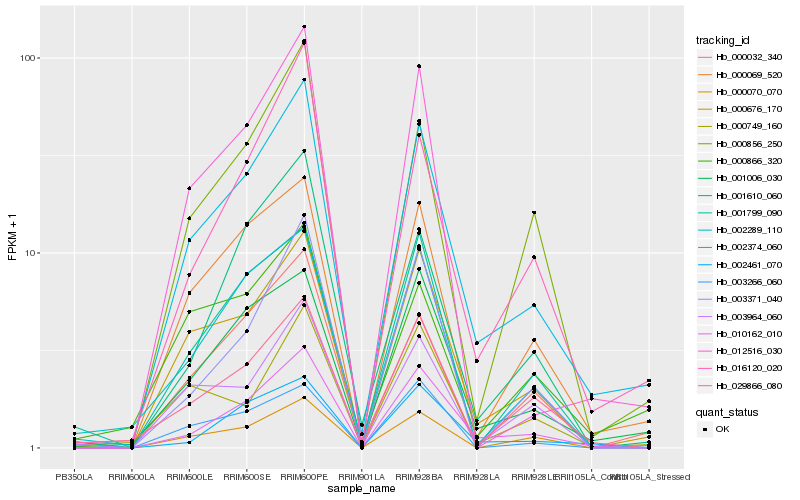
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_060 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_000676_170 |
0.0967116918 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 3 |
Hb_010162_010 |
0.1030926902 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 4 |
Hb_001006_030 |
0.1174117542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000069_520 |
0.1174649993 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 6 |
Hb_016120_020 |
0.1260910979 |
- |
- |
PREDICTED: tetrahydrocannabinolic acid synthase-like [Jatropha curcas] |
| 7 |
Hb_000856_250 |
0.135273116 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_012516_030 |
0.1410576109 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 9 |
Hb_000032_340 |
0.142499887 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 10 |
Hb_029866_080 |
0.1437024409 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 11 |
Hb_001610_060 |
0.1498053453 |
- |
- |
hypothetical protein TRIUR3_12067 [Triticum urartu] |
| 12 |
Hb_000070_070 |
0.1509190772 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 13 |
Hb_002289_110 |
0.1516214121 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 14 |
Hb_003964_060 |
0.1517568501 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |
| 15 |
Hb_000866_320 |
0.1551274927 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_003371_040 |
0.1608156669 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 17 |
Hb_001799_090 |
0.1620290756 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 18 |
Hb_003266_060 |
0.1629105695 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 19 |
Hb_000749_160 |
0.1645193532 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_002461_070 |
0.1656990275 |
- |
- |
gibberellin 2-oxidase, putative [Ricinus communis] |