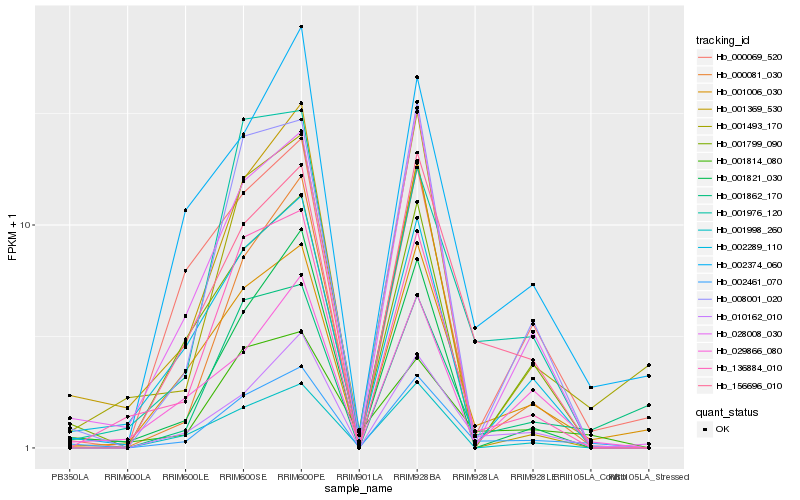
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002461_070 |
0.0 |
- |
- |
gibberellin 2-oxidase, putative [Ricinus communis] |
| 2 |
Hb_010162_010 |
0.1260081768 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 3 |
Hb_156696_010 |
0.146812329 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 4 |
Hb_001006_030 |
0.148148265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008001_020 |
0.1627715237 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 6 |
Hb_001821_030 |
0.1646112014 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 7 |
Hb_002374_060 |
0.1656990275 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_136884_010 |
0.1660469092 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_002289_110 |
0.1669355964 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 10 |
Hb_001814_080 |
0.1680685653 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 11 |
Hb_001799_090 |
0.169605706 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 12 |
Hb_028008_030 |
0.1722140716 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 13 |
Hb_000081_030 |
0.1773944032 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 14 |
Hb_029866_080 |
0.1811855021 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 15 |
Hb_001862_170 |
0.1826382389 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_001998_260 |
0.1833819718 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_000069_520 |
0.1873168287 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 18 |
Hb_001976_120 |
0.1888947564 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_001493_170 |
0.1889341901 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 20 |
Hb_001369_530 |
0.1909806858 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |