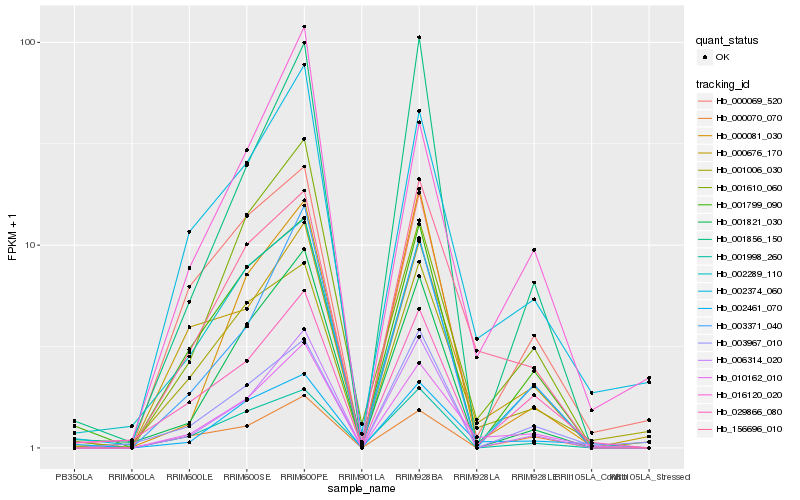
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010162_010 |
0.0 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 2 |
Hb_002374_060 |
0.1030926902 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_156696_010 |
0.1115022606 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 4 |
Hb_002461_070 |
0.1260081768 |
- |
- |
gibberellin 2-oxidase, putative [Ricinus communis] |
| 5 |
Hb_003371_040 |
0.135748096 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 6 |
Hb_006314_020 |
0.1404142077 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 7 |
Hb_001006_030 |
0.1406739131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000676_170 |
0.1438618409 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 9 |
Hb_029866_080 |
0.1457857215 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 10 |
Hb_016120_020 |
0.1547077463 |
- |
- |
PREDICTED: tetrahydrocannabinolic acid synthase-like [Jatropha curcas] |
| 11 |
Hb_001821_030 |
0.1597760408 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 12 |
Hb_000070_070 |
0.1626301667 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 13 |
Hb_001610_060 |
0.1634233986 |
- |
- |
hypothetical protein TRIUR3_12067 [Triticum urartu] |
| 14 |
Hb_002289_110 |
0.1646919212 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 15 |
Hb_001998_260 |
0.1649634717 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000069_520 |
0.1659793157 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 17 |
Hb_001799_090 |
0.1676307097 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 18 |
Hb_001856_150 |
0.1680157989 |
- |
- |
- |
| 19 |
Hb_000081_030 |
0.1690298145 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 20 |
Hb_003967_010 |
0.1709689916 |
- |
- |
cytochrome P450, putative [Ricinus communis] |