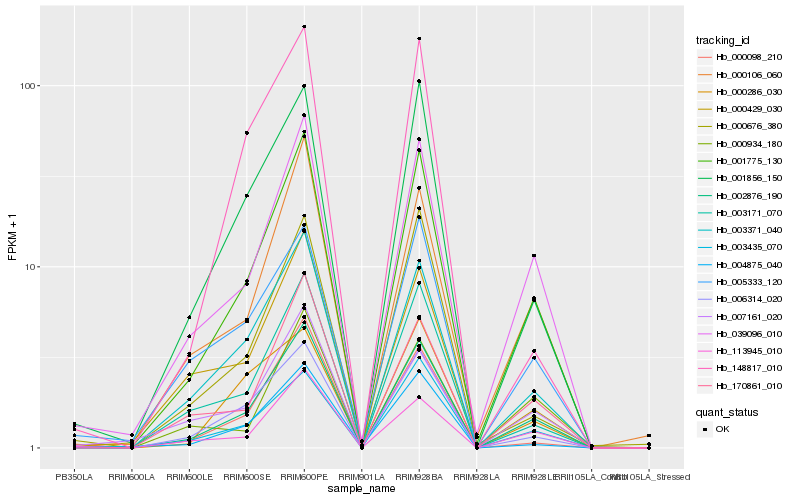
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_190 |
0.0 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 2 |
Hb_001775_130 |
0.0368000012 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 3 |
Hb_039096_010 |
0.0692768172 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_003371_040 |
0.0811809583 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 5 |
Hb_113945_010 |
0.1030672273 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000106_060 |
0.1049223912 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 7 |
Hb_006314_020 |
0.1057236398 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 8 |
Hb_001856_150 |
0.1074646524 |
- |
- |
- |
| 9 |
Hb_170861_010 |
0.1083899616 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003171_070 |
0.1103021979 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 11 |
Hb_003435_070 |
0.1158639873 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000098_210 |
0.1300291215 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 13 |
Hb_005333_120 |
0.1301837565 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 14 |
Hb_004875_040 |
0.1327509208 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 15 |
Hb_000934_180 |
0.134554959 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 16 |
Hb_000429_030 |
0.1364343374 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 17 |
Hb_007161_020 |
0.1409356225 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 18 |
Hb_000676_380 |
0.14287808 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 19 |
Hb_000286_030 |
0.1531450068 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_148817_010 |
0.1560594479 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |