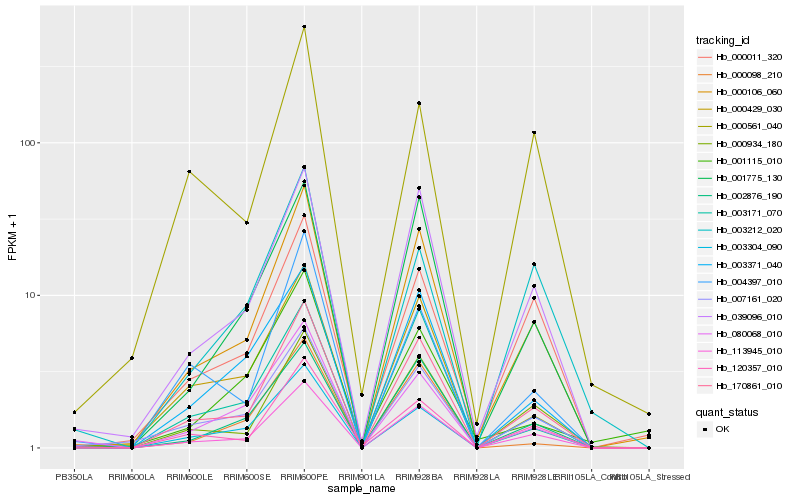
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 2 |
Hb_170861_010 |
0.0557700526 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 3 |
Hb_113945_010 |
0.06828117 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 4 |
Hb_039096_010 |
0.0847822399 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_000934_180 |
0.098474275 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 6 |
Hb_002876_190 |
0.1049223912 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 7 |
Hb_001775_130 |
0.1077389409 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 8 |
Hb_007161_020 |
0.1109945523 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 9 |
Hb_120357_010 |
0.122945615 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003371_040 |
0.1251038689 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 11 |
Hb_000011_320 |
0.1274855493 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 12 |
Hb_000429_030 |
0.1315085014 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 13 |
Hb_080068_010 |
0.1412248205 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 14 |
Hb_003171_070 |
0.1513215718 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 15 |
Hb_003304_090 |
0.1529650443 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 16 |
Hb_004397_010 |
0.1559125229 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000561_040 |
0.1597987748 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 18 |
Hb_001115_010 |
0.1617759668 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 2-like [Jatropha curcas] |
| 19 |
Hb_000098_210 |
0.1619942987 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 20 |
Hb_003212_020 |
0.1675799961 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |