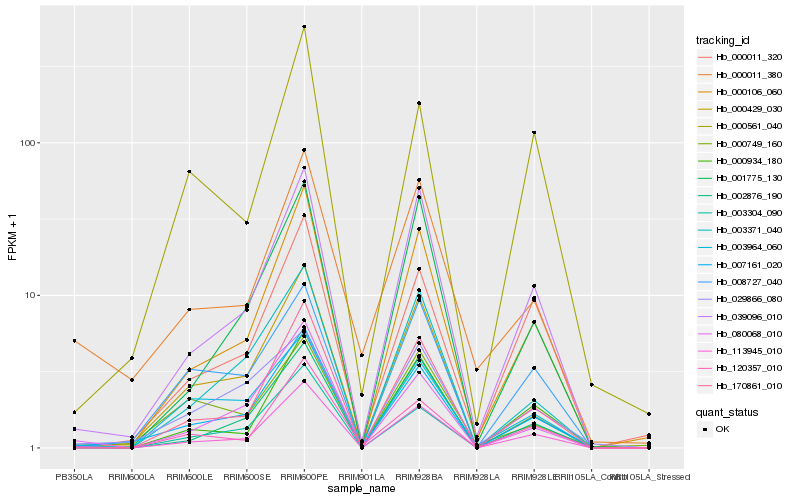
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007161_020 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 2 |
Hb_113945_010 |
0.0996469238 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 3 |
Hb_170861_010 |
0.1086730759 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000106_060 |
0.1109945523 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 5 |
Hb_000429_030 |
0.1140070476 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_039096_010 |
0.1225159263 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_080068_010 |
0.124967855 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 8 |
Hb_003371_040 |
0.1336507359 |
- |
- |
PREDICTED: uncharacterized protein LOC105650327 [Jatropha curcas] |
| 9 |
Hb_008727_040 |
0.1396085333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000934_180 |
0.1405981197 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 11 |
Hb_002876_190 |
0.1409356225 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 12 |
Hb_029866_080 |
0.1422388882 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 13 |
Hb_120357_010 |
0.1434326235 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000011_320 |
0.146561369 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 15 |
Hb_003964_060 |
0.1475206168 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |
| 16 |
Hb_003304_090 |
0.1477379607 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 17 |
Hb_000561_040 |
0.1485759053 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 18 |
Hb_001775_130 |
0.1550408688 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 19 |
Hb_000749_160 |
0.1563873602 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_000011_380 |
0.1566453905 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |