| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
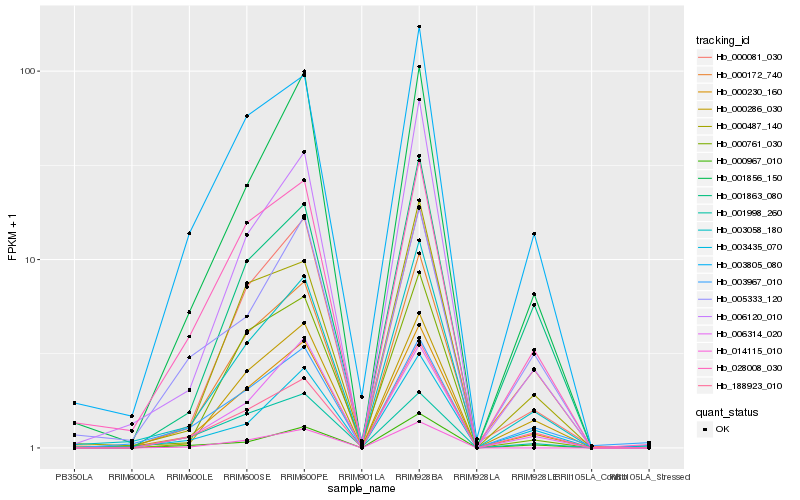
Hb_000230_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003058_180 |
0.0598821339 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 3 |
Hb_000081_030 |
0.0727014683 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 4 |
Hb_001856_150 |
0.0745275889 |
- |
- |
- |
| 5 |
Hb_000286_030 |
0.0840332411 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_028008_030 |
0.0858425329 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 7 |
Hb_006314_020 |
0.0917728015 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 8 |
Hb_003805_080 |
0.0931851989 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 9 |
Hb_188923_010 |
0.1060982616 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_006120_010 |
0.1071010871 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 11 |
Hb_003435_070 |
0.1108817126 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000761_030 |
0.1116506715 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 13 |
Hb_000172_740 |
0.1146073545 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 14 |
Hb_005333_120 |
0.1157160739 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 15 |
Hb_003967_010 |
0.1185978444 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000487_140 |
0.1196735456 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_014115_010 |
0.1225790777 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000967_010 |
0.1226434031 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 19 |
Hb_001998_260 |
0.1255736778 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_001863_080 |
0.1286188671 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |