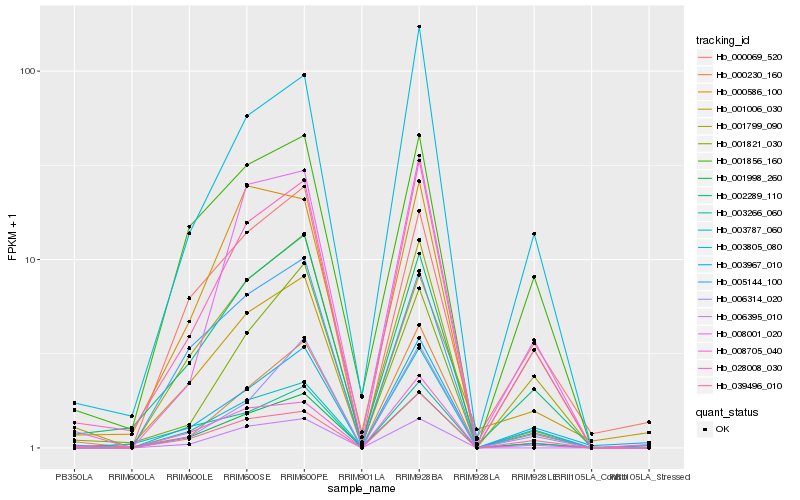
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001998_260 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_001799_090 |
0.0864347314 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 3 |
Hb_028008_030 |
0.0942933851 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 4 |
Hb_005144_100 |
0.1037430471 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 5 |
Hb_003266_060 |
0.1074838644 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 6 |
Hb_002289_110 |
0.1106472447 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 7 |
Hb_039496_010 |
0.1170367963 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 8 |
Hb_001856_160 |
0.1237063847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000230_160 |
0.1255736778 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006395_010 |
0.1259715099 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 11 |
Hb_001006_030 |
0.1277975504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008001_020 |
0.1289193891 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 13 |
Hb_003805_080 |
0.129972761 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 14 |
Hb_008705_040 |
0.1352974815 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 15 |
Hb_006314_020 |
0.1356362644 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 16 |
Hb_000069_520 |
0.1374277945 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 17 |
Hb_003967_010 |
0.1375731311 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_000586_100 |
0.1376624775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003787_060 |
0.1379409049 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 20 |
Hb_001821_030 |
0.1380021161 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |