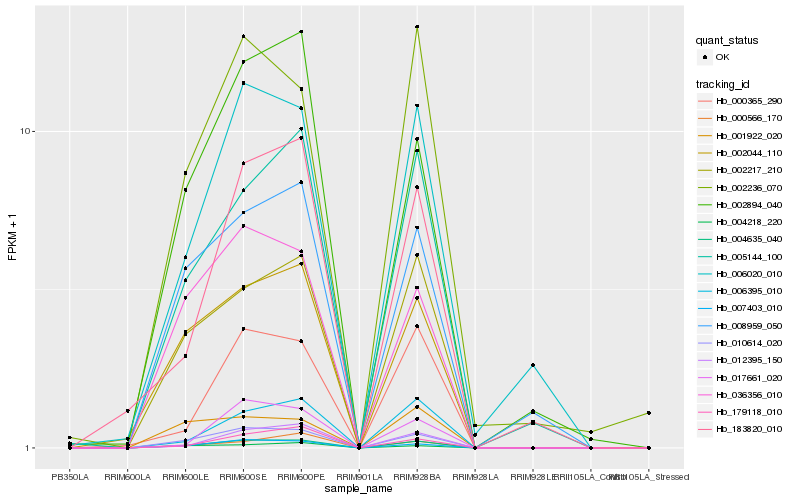
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004635_040 |
0.0 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 2 |
Hb_010614_020 |
0.0512750768 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 3 |
Hb_002217_210 |
0.0901174416 |
- |
- |
- |
| 4 |
Hb_007403_010 |
0.093343064 |
- |
- |
hypothetical protein JCGZ_11090 [Jatropha curcas] |
| 5 |
Hb_002044_110 |
0.1071745289 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 6 |
Hb_005144_100 |
0.1141248256 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_036356_010 |
0.1226104499 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_006020_010 |
0.1251796184 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 9 |
Hb_002894_040 |
0.1269612602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008959_050 |
0.1299186184 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_006395_010 |
0.1318905722 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 12 |
Hb_002236_070 |
0.1346688974 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 13 |
Hb_000365_290 |
0.1374574043 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_012395_150 |
0.140657378 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 15 |
Hb_183820_010 |
0.1491121169 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_001922_020 |
0.1552583776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004218_220 |
0.1564614636 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_179118_010 |
0.1584049896 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_000566_170 |
0.1607238311 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 20 |
Hb_017661_020 |
0.1609052066 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |