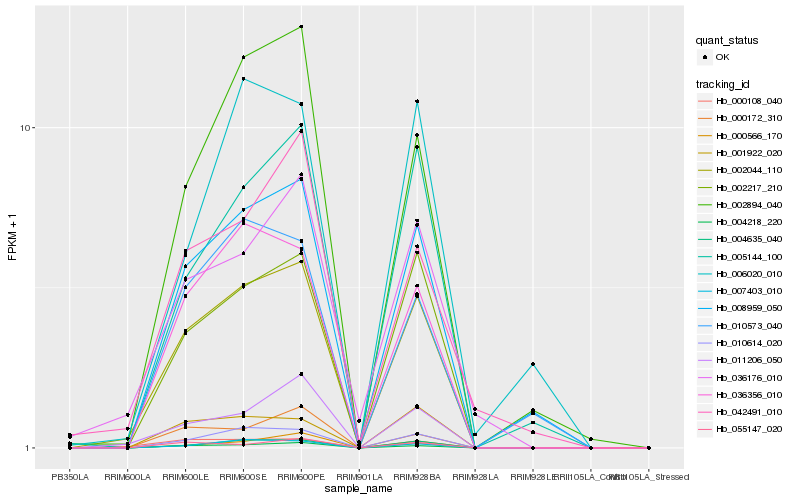
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002044_110 |
0.0 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 2 |
Hb_010614_020 |
0.0991783968 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 3 |
Hb_002217_210 |
0.1032285141 |
- |
- |
- |
| 4 |
Hb_004218_220 |
0.1071717798 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_004635_040 |
0.1071745289 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 6 |
Hb_036356_010 |
0.1151370161 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_008959_050 |
0.1152253669 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_011206_050 |
0.121436142 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 9 |
Hb_002894_040 |
0.1295943427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007403_010 |
0.1415944833 |
- |
- |
hypothetical protein JCGZ_11090 [Jatropha curcas] |
| 11 |
Hb_005144_100 |
0.1423867669 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 12 |
Hb_001922_020 |
0.1442017829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_036176_010 |
0.1569389534 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 14 |
Hb_010573_040 |
0.1613902337 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 15 |
Hb_042491_010 |
0.1634781381 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 16 |
Hb_000172_310 |
0.1656799819 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 17 |
Hb_000108_040 |
0.1657855448 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Gossypium raimondii] |
| 18 |
Hb_055147_020 |
0.1676162604 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 19 |
Hb_000566_170 |
0.1679599465 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 20 |
Hb_006020_010 |
0.1759611951 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |