| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
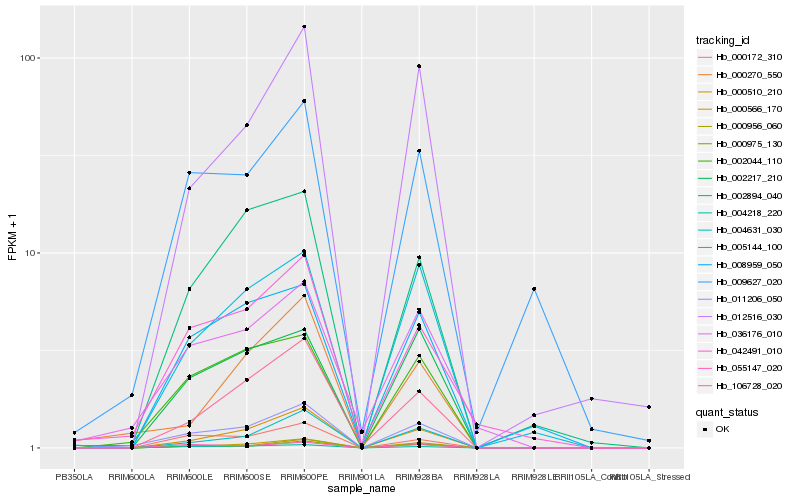
Hb_011206_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 2 |
Hb_000566_170 |
0.1147617716 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 3 |
Hb_002044_110 |
0.121436142 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 4 |
Hb_000510_210 |
0.1231334187 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_004218_220 |
0.1335367034 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000975_130 |
0.1428283127 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 7 |
Hb_000956_060 |
0.1434009426 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_042491_010 |
0.1456795652 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 9 |
Hb_106728_020 |
0.1459009046 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_036176_010 |
0.1466029458 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 11 |
Hb_004631_030 |
0.1482505723 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 12 |
Hb_012516_030 |
0.1541722492 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 13 |
Hb_000172_310 |
0.1561369959 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 14 |
Hb_005144_100 |
0.157494674 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 15 |
Hb_002217_210 |
0.1614178512 |
- |
- |
- |
| 16 |
Hb_055147_020 |
0.1645434766 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 17 |
Hb_002894_040 |
0.1657146156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009627_020 |
0.1689014077 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 19 |
Hb_000270_550 |
0.1715910228 |
- |
- |
- |
| 20 |
Hb_008959_050 |
0.1734379152 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |