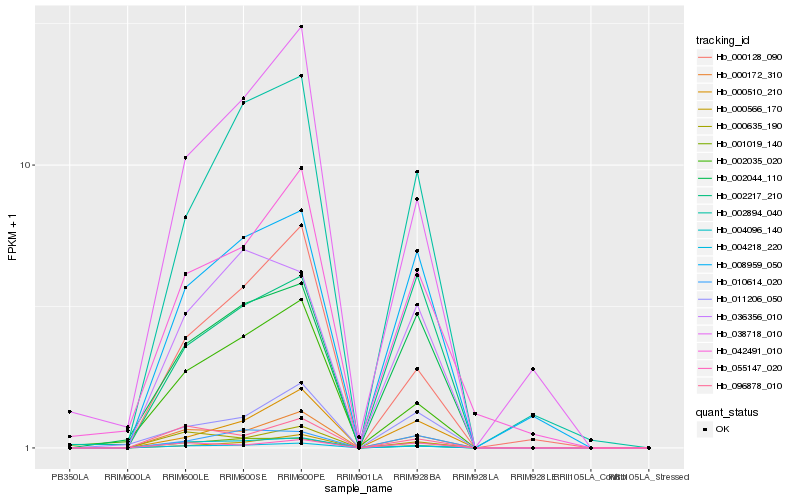
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_220 |
0.0 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000172_310 |
0.0722051274 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 3 |
Hb_002044_110 |
0.1071717798 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 4 |
Hb_002894_040 |
0.1172738093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008959_050 |
0.1246273469 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_055147_020 |
0.1320560409 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 7 |
Hb_011206_050 |
0.1335367034 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 8 |
Hb_000128_090 |
0.135343193 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 9 |
Hb_036356_010 |
0.1383669139 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_004096_140 |
0.1398923658 |
- |
- |
Lipase, putative [Ricinus communis] |
| 11 |
Hb_010614_020 |
0.1399140498 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 12 |
Hb_000566_170 |
0.1403832735 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 13 |
Hb_042491_010 |
0.1408964705 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 14 |
Hb_001019_140 |
0.1427675912 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_038718_010 |
0.1427799666 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_096878_010 |
0.1445847743 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 17 |
Hb_002035_020 |
0.1460372802 |
transcription factor |
TF Family: TRAF |
hypothetical protein B456_011G050200 [Gossypium raimondii] |
| 18 |
Hb_000510_210 |
0.148191677 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000635_190 |
0.1503144971 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 20 |
Hb_002217_210 |
0.1520921373 |
- |
- |
- |