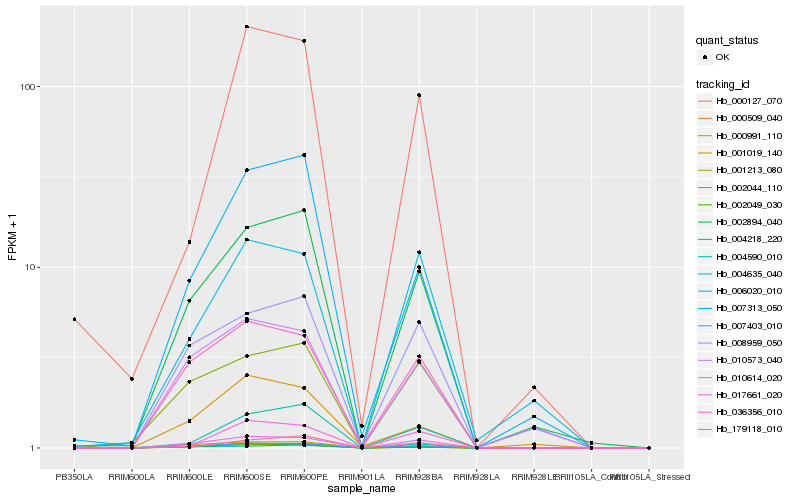
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007403_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_11090 [Jatropha curcas] |
| 2 |
Hb_010614_020 |
0.0625623718 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 3 |
Hb_004635_040 |
0.093343064 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 4 |
Hb_002894_040 |
0.1036485909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_036356_010 |
0.1038699966 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_002044_110 |
0.1415944833 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 7 |
Hb_017661_020 |
0.144043274 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 8 |
Hb_004590_010 |
0.1447024876 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 9 |
Hb_007313_050 |
0.1504342759 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 10 |
Hb_010573_040 |
0.1517177773 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 11 |
Hb_000509_040 |
0.1518949222 |
- |
- |
hypothetical protein VITISV_009716 [Vitis vinifera] |
| 12 |
Hb_002049_030 |
0.1540678942 |
- |
- |
PREDICTED: uncharacterized protein LOC105635940 isoform X3 [Jatropha curcas] |
| 13 |
Hb_006020_010 |
0.1540772999 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 14 |
Hb_179118_010 |
0.1542204439 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_000991_110 |
0.1547096255 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 16 |
Hb_001213_080 |
0.1556980452 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 17 |
Hb_000127_070 |
0.1563528657 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 18 |
Hb_008959_050 |
0.1576001467 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001019_140 |
0.1602879413 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_004218_220 |
0.1624284706 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |