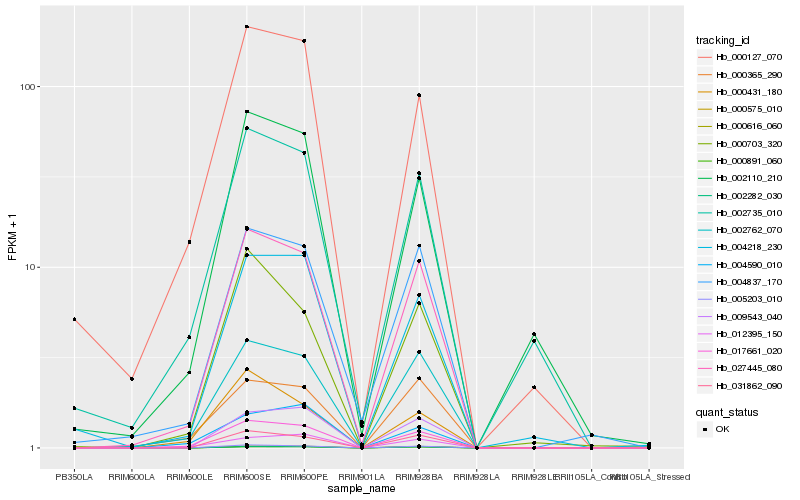
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017661_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 2 |
Hb_027445_080 |
0.0670177072 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 3 |
Hb_002762_070 |
0.0836718229 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 4 |
Hb_000127_070 |
0.1020927564 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 5 |
Hb_012395_150 |
0.1049800834 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 6 |
Hb_005203_010 |
0.1115202482 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 7 |
Hb_004218_230 |
0.1208696044 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 8 |
Hb_004590_010 |
0.1215834134 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 9 |
Hb_000431_180 |
0.122084739 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 10 |
Hb_000365_290 |
0.1233935018 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002735_010 |
0.1249414418 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 12 |
Hb_004837_170 |
0.1264429987 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 13 |
Hb_002110_210 |
0.1284100418 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 14 |
Hb_002282_030 |
0.1287416368 |
- |
- |
- |
| 15 |
Hb_000891_060 |
0.129089028 |
- |
- |
hypothetical protein JCGZ_11234 [Jatropha curcas] |
| 16 |
Hb_000575_010 |
0.1291922544 |
- |
- |
PREDICTED: uncharacterized protein LOC103454030 [Malus domestica] |
| 17 |
Hb_000616_060 |
0.129533338 |
- |
- |
- |
| 18 |
Hb_031862_090 |
0.1308531931 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 19 |
Hb_009543_040 |
0.132805774 |
- |
- |
- |
| 20 |
Hb_000703_320 |
0.1371505015 |
- |
- |
receptor-kinase, putative [Ricinus communis] |