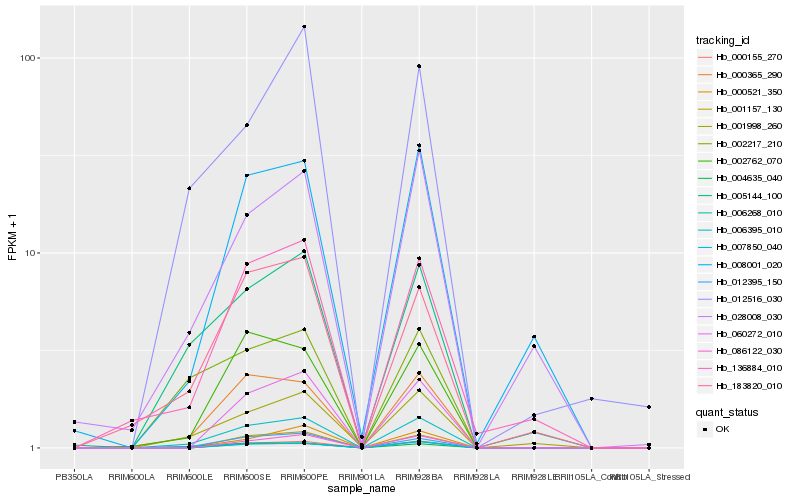
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006395_010 |
0.0 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 2 |
Hb_000365_290 |
0.0984642777 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_012395_150 |
0.0993166161 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 4 |
Hb_001998_260 |
0.1259715099 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_005144_100 |
0.1267376351 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_004635_040 |
0.1318905722 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 7 |
Hb_002762_070 |
0.1347602546 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 8 |
Hb_000155_270 |
0.1401098705 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 9 |
Hb_000521_350 |
0.1418380139 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_183820_010 |
0.143668824 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 11 |
Hb_060272_010 |
0.1437654221 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 12 |
Hb_136884_010 |
0.1469141019 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 13 |
Hb_002217_210 |
0.1480008458 |
- |
- |
- |
| 14 |
Hb_001157_130 |
0.1488285588 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 15 |
Hb_086122_030 |
0.150764056 |
- |
- |
D-glycerate 3-kinase, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_006268_010 |
0.1520105903 |
- |
- |
hypothetical protein EUGRSUZ_B00966 [Eucalyptus grandis] |
| 17 |
Hb_008001_020 |
0.1522636055 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 18 |
Hb_012516_030 |
0.1553574388 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 2 {ECO:0000303 |
| 19 |
Hb_007850_040 |
0.1558441732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_028008_030 |
0.1563131362 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |