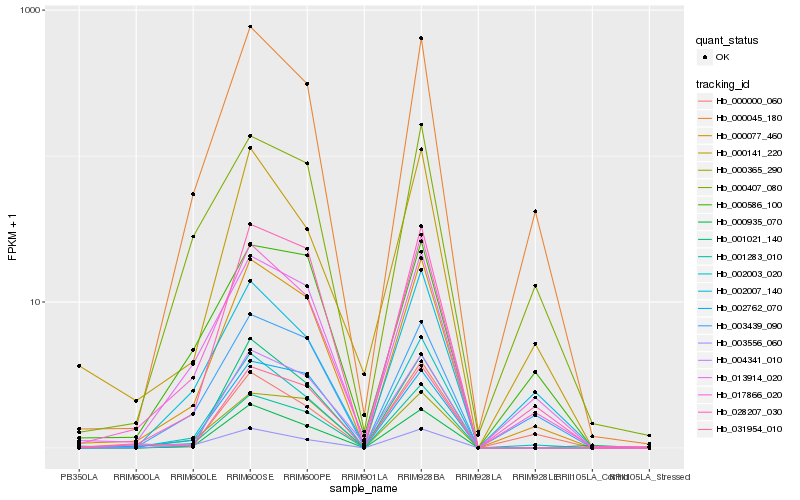
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_460 |
0.0 |
- |
- |
- |
| 2 |
Hb_028207_030 |
0.0739890702 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 3 |
Hb_017866_020 |
0.0779337302 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 4 |
Hb_000045_180 |
0.0828106675 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 5 |
Hb_001283_010 |
0.0913219446 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 6 |
Hb_013914_020 |
0.0934456859 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 7 |
Hb_002003_020 |
0.1052544578 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 8 |
Hb_004341_010 |
0.1087685529 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 9 |
Hb_031954_010 |
0.1100707366 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_003439_090 |
0.1108111941 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_003556_060 |
0.1174885138 |
- |
- |
- |
| 12 |
Hb_000365_290 |
0.1177930103 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002762_070 |
0.118122615 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 14 |
Hb_000407_080 |
0.1291409873 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 15 |
Hb_000935_070 |
0.1310766414 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001021_140 |
0.1325600545 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 17 |
Hb_000000_060 |
0.1330040366 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 18 |
Hb_000141_220 |
0.1331566722 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 19 |
Hb_000586_100 |
0.1358094359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002007_140 |
0.1368110792 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |