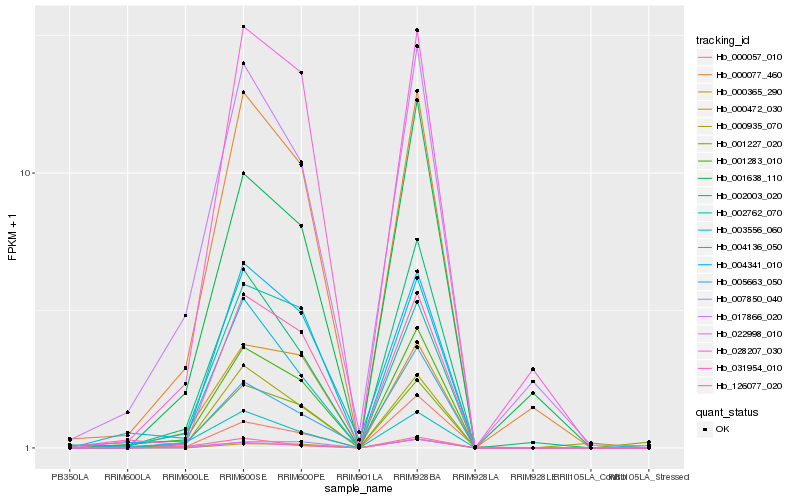
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001283_010 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 2 |
Hb_000077_460 |
0.0913219446 |
- |
- |
- |
| 3 |
Hb_004136_050 |
0.0943049055 |
- |
- |
- |
| 4 |
Hb_031954_010 |
0.0949853152 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_017866_020 |
0.1068016701 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 6 |
Hb_002003_020 |
0.1079508694 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 7 |
Hb_004341_010 |
0.1112816147 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 8 |
Hb_000365_290 |
0.111554135 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003556_060 |
0.1222986884 |
- |
- |
- |
| 10 |
Hb_028207_030 |
0.1233542241 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 11 |
Hb_002762_070 |
0.1300807724 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 12 |
Hb_005663_050 |
0.132620051 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_001227_020 |
0.1357318896 |
- |
- |
- |
| 14 |
Hb_001638_110 |
0.1375069536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000935_070 |
0.1409224402 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000057_010 |
0.1445391058 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 17 |
Hb_022998_010 |
0.1463351572 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_126077_020 |
0.1468663397 |
- |
- |
Kinase family protein with leucine-rich repeat domain [Theobroma cacao] |
| 19 |
Hb_000472_030 |
0.1480641466 |
- |
- |
- |
| 20 |
Hb_007850_040 |
0.1481293432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |