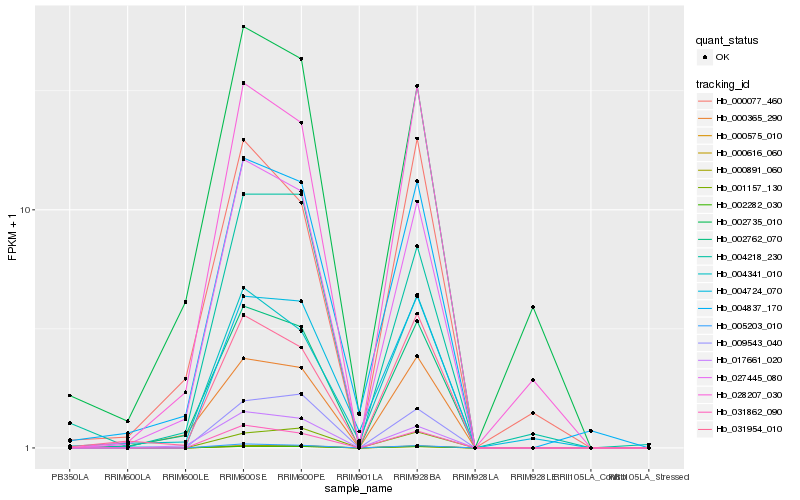
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 2 |
Hb_027445_080 |
0.0874089986 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 3 |
Hb_004341_010 |
0.091428456 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 4 |
Hb_002762_070 |
0.0952081206 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 5 |
Hb_031954_010 |
0.1119923133 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_004724_070 |
0.1199683219 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002282_030 |
0.1241481278 |
- |
- |
- |
| 8 |
Hb_000365_290 |
0.1243604698 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000891_060 |
0.1261476722 |
- |
- |
hypothetical protein JCGZ_11234 [Jatropha curcas] |
| 10 |
Hb_017661_020 |
0.1264429987 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 11 |
Hb_031862_090 |
0.126758201 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 12 |
Hb_000616_060 |
0.1275902979 |
- |
- |
- |
| 13 |
Hb_000575_010 |
0.1284519908 |
- |
- |
PREDICTED: uncharacterized protein LOC103454030 [Malus domestica] |
| 14 |
Hb_005203_010 |
0.1319249189 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 15 |
Hb_028207_030 |
0.1329547167 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 16 |
Hb_002735_010 |
0.1437721768 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 44 [Prunus mume] |
| 17 |
Hb_004218_230 |
0.1439861545 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 18 |
Hb_009543_040 |
0.1460101303 |
- |
- |
- |
| 19 |
Hb_000077_460 |
0.1543957593 |
- |
- |
- |
| 20 |
Hb_001157_130 |
0.1562889932 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |