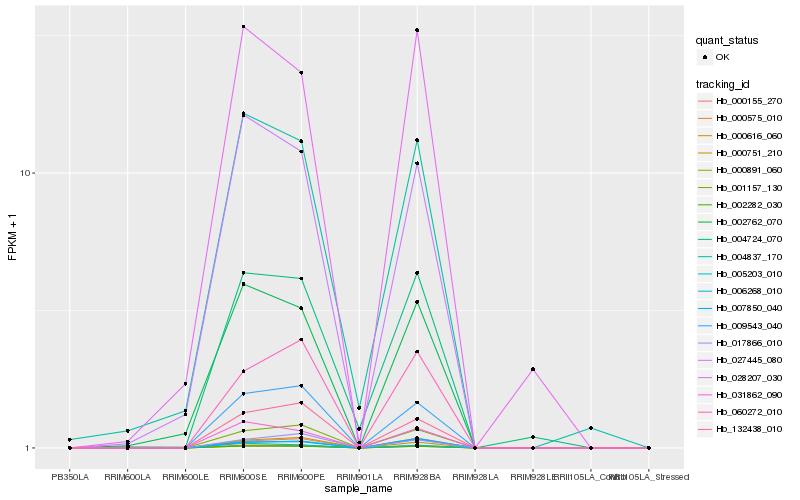
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002282_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000891_060 |
0.0081229278 |
- |
- |
hypothetical protein JCGZ_11234 [Jatropha curcas] |
| 3 |
Hb_000616_060 |
0.0130473353 |
- |
- |
- |
| 4 |
Hb_000575_010 |
0.0162285038 |
- |
- |
PREDICTED: uncharacterized protein LOC103454030 [Malus domestica] |
| 5 |
Hb_009543_040 |
0.0588140304 |
- |
- |
- |
| 6 |
Hb_001157_130 |
0.0694399965 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 7 |
Hb_031862_090 |
0.0714975388 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 8 |
Hb_005203_010 |
0.0792153768 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 9 |
Hb_132438_010 |
0.0835253315 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 10 |
Hb_000155_270 |
0.0910227572 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 11 |
Hb_027445_080 |
0.0941703363 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 12 |
Hb_000751_210 |
0.0980704113 |
- |
- |
PREDICTED: petal death protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_007850_040 |
0.0988029587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_060272_010 |
0.1024721447 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 15 |
Hb_002762_070 |
0.1042780316 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 16 |
Hb_004724_070 |
0.1222036306 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_017866_010 |
0.1223148265 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 18 |
Hb_028207_030 |
0.123645059 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 19 |
Hb_004837_170 |
0.1241481278 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 20 |
Hb_006268_010 |
0.124437324 |
- |
- |
hypothetical protein EUGRSUZ_B00966 [Eucalyptus grandis] |