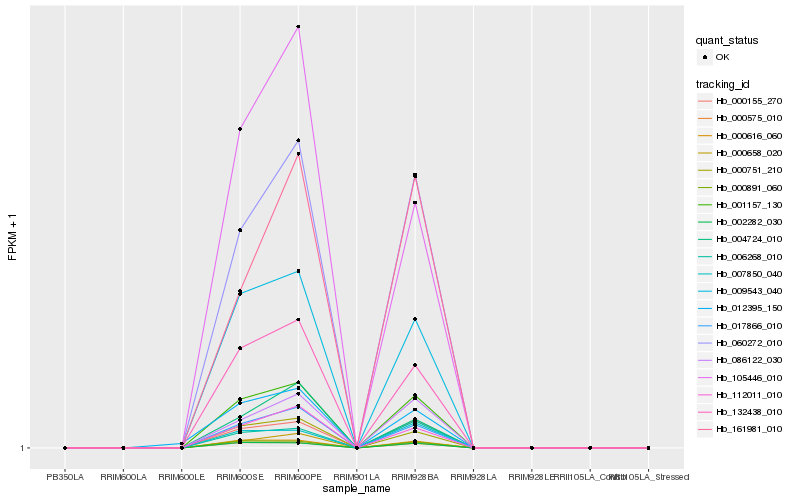
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001157_130 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 2 |
Hb_060272_010 |
0.0397489983 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 3 |
Hb_009543_040 |
0.0497124906 |
- |
- |
- |
| 4 |
Hb_132438_010 |
0.0537687525 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 5 |
Hb_000575_010 |
0.0564574029 |
- |
- |
PREDICTED: uncharacterized protein LOC103454030 [Malus domestica] |
| 6 |
Hb_000616_060 |
0.0582662716 |
- |
- |
- |
| 7 |
Hb_000155_270 |
0.0584631651 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 8 |
Hb_000891_060 |
0.0623945224 |
- |
- |
hypothetical protein JCGZ_11234 [Jatropha curcas] |
| 9 |
Hb_017866_010 |
0.0660283742 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 10 |
Hb_002282_030 |
0.0694399965 |
- |
- |
- |
| 11 |
Hb_000751_210 |
0.0718085931 |
- |
- |
PREDICTED: petal death protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_086122_030 |
0.0855511751 |
- |
- |
D-glycerate 3-kinase, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_112011_010 |
0.0972384085 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_105446_010 |
0.1044908302 |
- |
- |
- |
| 15 |
Hb_007850_040 |
0.1067986555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_161981_010 |
0.1071756101 |
- |
- |
brassinosteroid insensitive 1 -like protein [Gossypium arboreum] |
| 17 |
Hb_006268_010 |
0.10961119 |
- |
- |
hypothetical protein EUGRSUZ_B00966 [Eucalyptus grandis] |
| 18 |
Hb_000658_020 |
0.1187791006 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_004724_010 |
0.1212223227 |
- |
- |
- |
| 20 |
Hb_012395_150 |
0.1278024464 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |